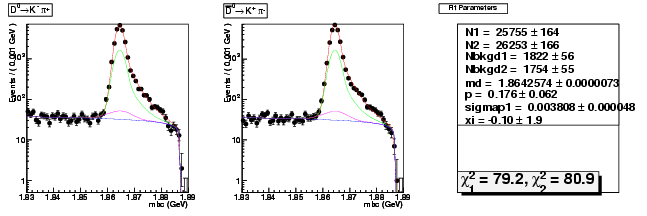
Mode 0 : D0 &rarr K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 25755.4209228000 | 164.4685751991 |
| N2 | 26253.2462201141 | 165.9284500250 |
| Nbkgd1 | 1821.6789322138 | 55.8253472669 |
| Nbkgd2 | 1753.8497607670 | 55.0686117774 |
| md | 1.8642574075 | 0.0000073402 |
| p | 0.1759241342 | 0.0624577407 |
| sigmap1 | 0.0038075474 | 0.0000483909 |
| xi | -0.1000087095 | 1.9207990283 |
| chisq1 | 79.2174157065 | 0.0000000000 |
| chisq2 | 80.8877354625 | 0.0000000000 |
dhad fit faplus -t d -m 0
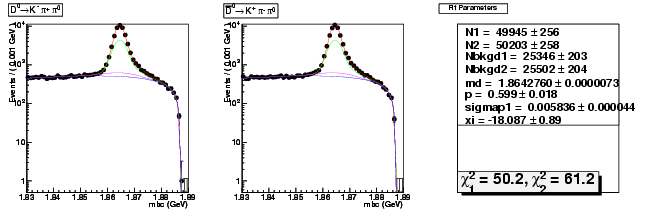
Mode 1 : D0 &rarr K- &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 49944.5882143617 | 256.3266536252 |
| N2 | 50203.2064052084 | 257.5967823906 |
| Nbkgd1 | 25345.5617519814 | 202.7408891469 |
| Nbkgd2 | 25501.9065860905 | 204.0955210210 |
| md | 1.8642759989 | 0.0000072961 |
| p | 0.5992741389 | 0.0180170976 |
| sigmap1 | 0.0058356034 | 0.0000440795 |
| xi | -18.0868238019 | 0.8890990107 |
| chisq1 | 50.2362034455 | 0.0000000000 |
| chisq2 | 61.2468619320 | 0.0000000000 |
dhad fit faplus -t d -m 1
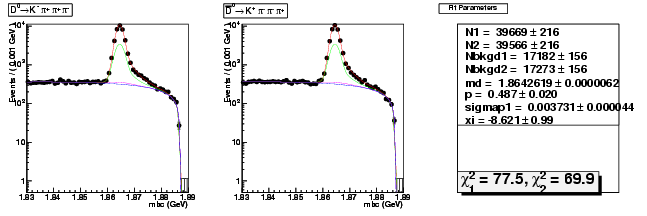
Mode 3 : D0 &rarr K- &pi+ &pi- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 39668.5637307559 | 216.1168313528 |
| N2 | 39566.2773868127 | 215.8398135461 |
| Nbkgd1 | 17182.2656148671 | 155.6307380382 |
| Nbkgd2 | 17272.5262232072 | 155.8624593654 |
| md | 1.8642619154 | 0.0000061834 |
| p | 0.4865708420 | 0.0203699665 |
| sigmap1 | 0.0037314433 | 0.0000438218 |
| xi | -8.6205955452 | 0.9940010551 |
| chisq1 | 77.4523823887 | 0.0000000000 |
| chisq2 | 69.9144230296 | 0.0000000000 |
dhad fit faplus -t d -m 3
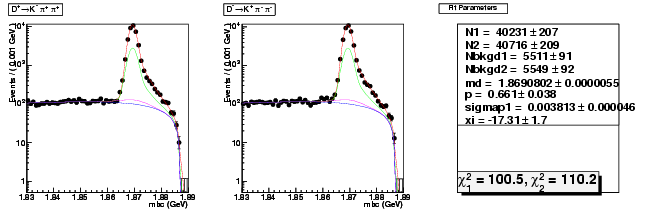
Mode 200 : D+ &rarr K- &pi+ &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 40230.5979532126 | 207.3437545738 |
| N2 | 40715.8149636397 | 208.6494485131 |
| Nbkgd1 | 5510.5579756565 | 90.9723067394 |
| Nbkgd2 | 5549.2927899542 | 91.5028263979 |
| md | 1.8690802275 | 0.0000055349 |
| p | 0.6606867861 | 0.0377378236 |
| sigmap1 | 0.0038133465 | 0.0000455674 |
| xi | -17.3143764407 | 1.6799849902 |
| chisq1 | 100.4526722519 | 0.0000000000 |
| chisq2 | 110.1790156451 | 0.0000000000 |
dhad fit faplus -t d -m 200
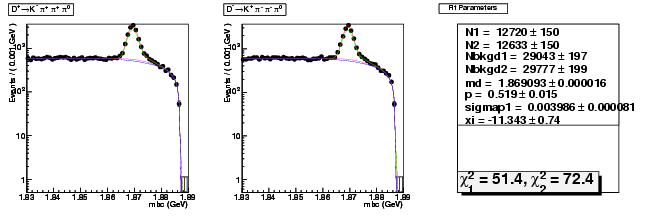
Mode 201 : D+ &rarr K- &pi+ &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 12719.8033760702 | 149.5056733758 |
| N2 | 12633.3512457598 | 149.5028679079 |
| Nbkgd1 | 29043.2522019477 | 196.6583527875 |
| Nbkgd2 | 29776.7479747678 | 198.7318818029 |
| md | 1.8690929207 | 0.0000156710 |
| p | 0.5186707668 | 0.0150488694 |
| sigmap1 | 0.0039856649 | 0.0000806575 |
| xi | -11.3425857313 | 0.7428721746 |
| chisq1 | 51.4308740573 | 0.0000000000 |
| chisq2 | 72.4167404760 | 0.0000000000 |
dhad fit faplus -t d -m 201
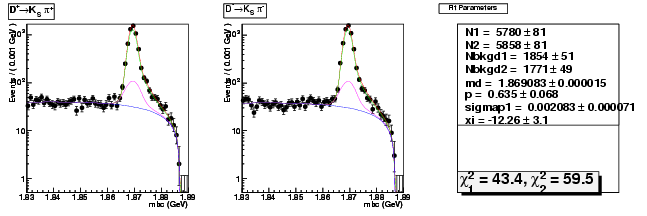
Mode 202 : D+ &rarr KS0 &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 5779.5008453805 | 80.7115974778 |
| N2 | 5858.4498964321 | 80.8506121290 |
| Nbkgd1 | 1853.5048095139 | 50.8792358029 |
| Nbkgd2 | 1770.5427309627 | 49.4896705468 |
| md | 1.8690831261 | 0.0000150781 |
| p | 0.6345729614 | 0.0681211150 |
| sigmap1 | 0.0020827607 | 0.0000712725 |
| xi | -12.2636833295 | 3.0971932023 |
| chisq1 | 43.3696562532 | 0.0000000000 |
| chisq2 | 59.4599141369 | 0.0000000000 |
dhad fit faplus -t d -m 202
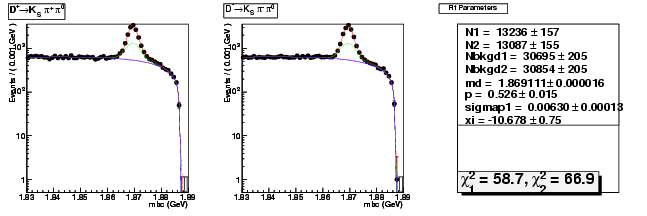
Mode 203 : D+ &rarr KS0 &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 13236.4560089292 | 157.0155578454 |
| N2 | 13087.2633723359 | 155.3295451818 |
| Nbkgd1 | 30694.9816633595 | 205.2087197272 |
| Nbkgd2 | 30854.1599839012 | 204.6740828840 |
| md | 1.8691109494 | 0.0000161317 |
| p | 0.5255711655 | 0.0150556146 |
| sigmap1 | 0.0062988149 | 0.0001304658 |
| xi | -10.6779725546 | 0.7456037560 |
| chisq1 | 58.6676288854 | 0.0000000000 |
| chisq2 | 66.8528226707 | 0.0000000000 |
dhad fit faplus -t d -m 203
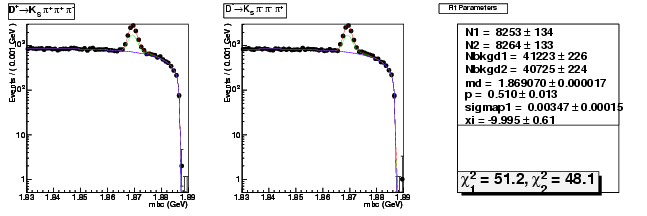
Mode 204 : D+ &rarr KS0 &pi+ &pi+ &pi-
| Name | Value | Error |
|---|---|---|
| N1 | 8253.2160802297 | 133.7608806734 |
| N2 | 8263.6467363641 | 133.2323166914 |
| Nbkgd1 | 41222.9884607750 | 225.5198973686 |
| Nbkgd2 | 40724.6120872246 | 224.0748160261 |
| md | 1.8690698222 | 0.0000170875 |
| p | 0.5095619821 | 0.0125263725 |
| sigmap1 | 0.0034670051 | 0.0001539196 |
| xi | -9.9952571938 | 0.6060411725 |
| chisq1 | 51.2421717196 | 0.0000000000 |
| chisq2 | 48.1159100958 | 0.0000000000 |
dhad fit faplus -t d -m 204
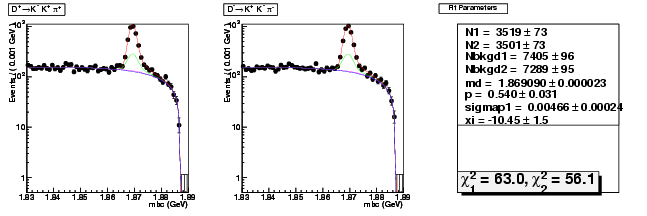
Mode 205 : D+ &rarr K+ K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 3519.2728003737 | 73.1978770945 |
| N2 | 3500.6694923084 | 72.6486036503 |
| Nbkgd1 | 7405.0052333987 | 96.1433767929 |
| Nbkgd2 | 7288.5898935129 | 95.2136966580 |
| md | 1.8690903853 | 0.0000234010 |
| p | 0.5404001776 | 0.0305437052 |
| sigmap1 | 0.0046648589 | 0.0002374219 |
| xi | -10.4546173140 | 1.4597436576 |
| chisq1 | 63.0038796109 | 0.0000000000 |
| chisq2 | 56.1042426164 | 0.0000000000 |
dhad fit faplus -t d -m 205