
 |
 |
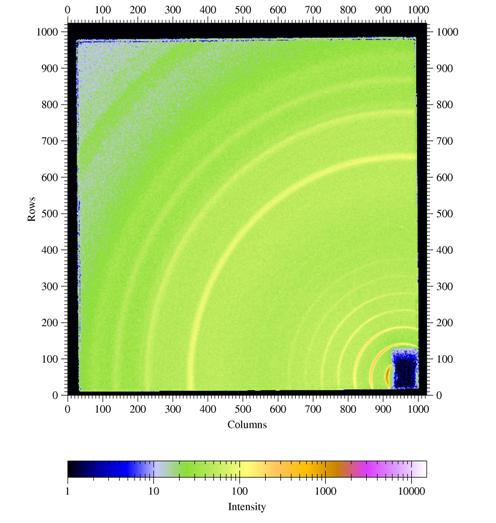
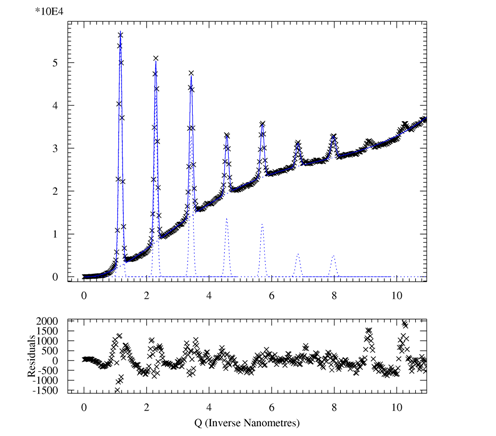
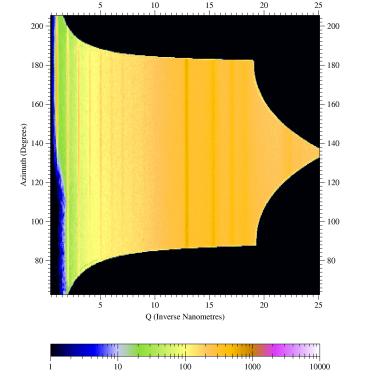
| Fig 5a: Radial scan (90-890 pixels in 800
steps), integrated over azimuth range of 90°-120°. The (00L) reflections indicative of lamellae formed by the ordering of the alkyl side-chains show up prodominantly in the perpendicular direction, i.e. the lamellae are parallel to the surface. The broad powder ring is due to the amorphous silicon oxide layer at the wafer surface. |
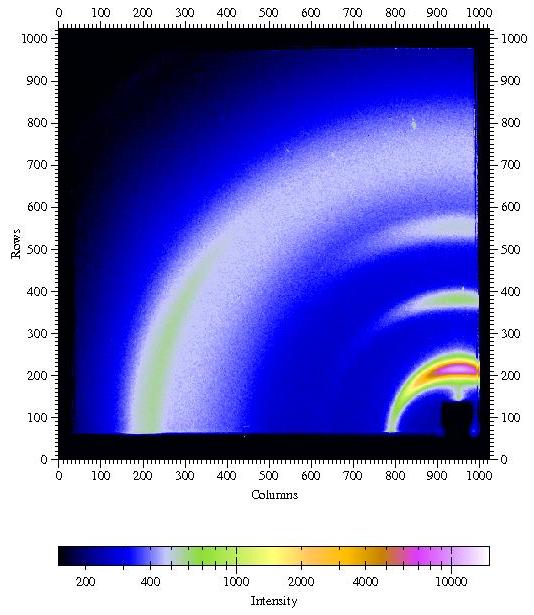
Fig 5b. Radial scan (90-890 pixels in 800
steps), integrated over azimuth range of 120°-180°. In the in-plane direction the (100) peak due to the pi-stacking of the aromatic rings of the polymer back-bone can be found, i.e. polymer chains are parallel to the surface and parallel to each other. Weak (00L) powderrings indicate that the lamellae are not perfectly parallel to the substrate. |
 |
 |
 |
 |