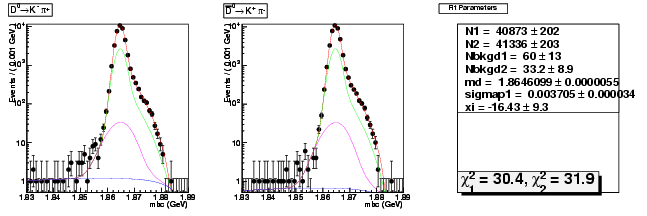
Mode 0 : D0 &rarr K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 40873.4995656324 | 202.4507211987 |
| N2 | 41335.8816466112 | 203.4272773660 |
| Nbkgd1 | 59.6456684146 | 13.1213789000 |
| Nbkgd2 | 33.2299010683 | 8.9054422736 |
| md | 1.8646099463 | 0.0000055458 |
| p | 0.5000000000 | 0.0000000000 |
| sigmap1 | 0.0037049897 | 0.0000342921 |
| xi | -16.4297488606 | 9.2655229165 |
| chisq1 | 30.4260214414 | 0.0000000000 |
| chisq2 | 31.8956404338 | 0.0000000000 |
dhad fit faplus -t s -m 0
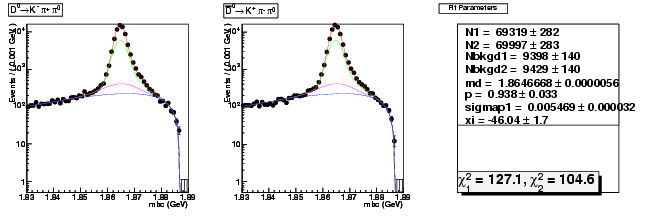
Mode 1 : D0 &rarr K- &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 69318.8084772648 | 281.7564058697 |
| N2 | 69996.9492734739 | 283.2889565374 |
| Nbkgd1 | 9398.3704602482 | 139.5192272692 |
| Nbkgd2 | 9429.1832845818 | 140.3113578813 |
| md | 1.8646667629 | 0.0000056424 |
| p | 0.9380774064 | 0.0333081644 |
| sigmap1 | 0.0054686209 | 0.0000318101 |
| xi | -46.0416764720 | 1.6724869174 |
| chisq1 | 127.0755772805 | 0.0000000000 |
| chisq2 | 104.5869676423 | 0.0000000000 |
dhad fit faplus -t s -m 1
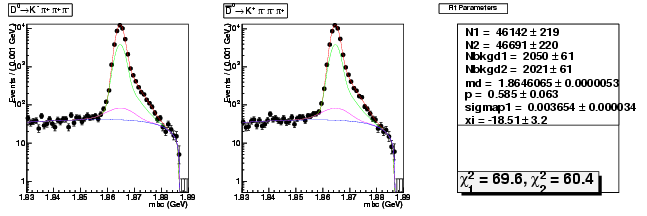
Mode 3 : D0 &rarr K- &pi+ &pi- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 46142.1668773504 | 218.7015887068 |
| N2 | 46691.0275897158 | 219.8948234426 |
| Nbkgd1 | 2049.8618310997 | 61.1468694819 |
| Nbkgd2 | 2021.0089756444 | 60.7024196808 |
| md | 1.8646065231 | 0.0000053006 |
| p | 0.5850538142 | 0.0626452889 |
| sigmap1 | 0.0036543993 | 0.0000342313 |
| xi | -18.5096670275 | 3.2353098477 |
| chisq1 | 69.5554413769 | 0.0000000000 |
| chisq2 | 60.3724068921 | 0.0000000000 |
dhad fit faplus -t s -m 3
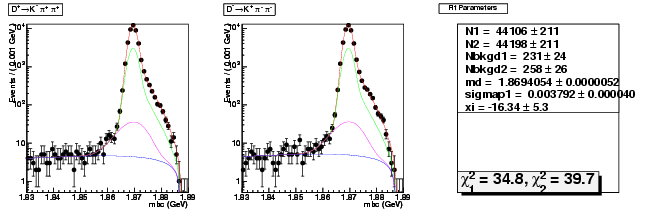
Mode 200 : D+ &rarr K- &pi+ &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 44106.2723022588 | 210.8720743927 |
| N2 | 44197.6949530812 | 211.1796621166 |
| Nbkgd1 | 230.9333224547 | 24.3032583159 |
| Nbkgd2 | 257.5221049606 | 25.6493986288 |
| md | 1.8694053815 | 0.0000051661 |
| p | 0.5000000000 | 0.0000000000 |
| sigmap1 | 0.0037918489 | 0.0000397375 |
| xi | -16.3428507478 | 5.2704380023 |
| chisq1 | 34.8367767085 | 0.0000000000 |
| chisq2 | 39.7011091932 | 0.0000000000 |
dhad fit faplus -t s -m 200
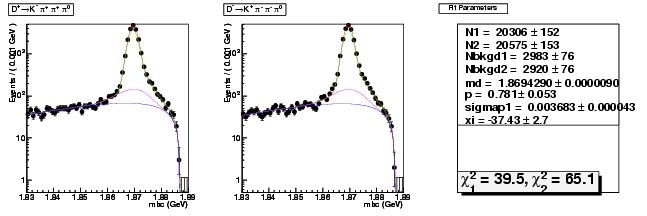
Mode 201 : D+ &rarr K- &pi+ &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 20306.2044037420 | 151.8110972247 |
| N2 | 20574.7750785515 | 152.8421968590 |
| Nbkgd1 | 2983.0349943914 | 75.6933735532 |
| Nbkgd2 | 2920.4330089231 | 75.5812968480 |
| md | 1.8694289999 | 0.0000090148 |
| p | 0.7811830849 | 0.0525439422 |
| sigmap1 | 0.0036834269 | 0.0000432917 |
| xi | -37.4308574995 | 2.7101129736 |
| chisq1 | 39.5047278474 | 0.0000000000 |
| chisq2 | 65.0807651550 | 0.0000000000 |
dhad fit faplus -t s -m 201
Mode 202 : D+ &rarr KS0 &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 36654.0077167342 | 192.0632036410 |
| N2 | 36725.3219145314 | 192.2557456192 |
| Nbkgd1 | 128.2606687770 | 19.0319750466 |
| Nbkgd2 | 125.9561668982 | 19.0564576382 |
| md | 1.8694200463 | 0.0000055986 |
| p | 0.5000000000 | 0.0000000000 |
| sigmap1 | 0.0020000000 | 0.0000006376 |
| xi | -30.0754806703 | 7.2457249588 |
| chisq1 | 84.2897802386 | 0.0000000000 |
| chisq2 | 63.8468864915 | 0.0000000000 |
dhad fit faplus -t s -m 202
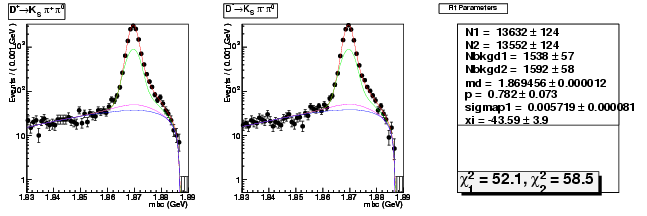
Mode 203 : D+ &rarr KS0 &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 13631.6902162254 | 123.8726771518 |
| N2 | 13551.5767779127 | 123.7341595469 |
| Nbkgd1 | 1538.2204991423 | 57.0214918263 |
| Nbkgd2 | 1592.1171469270 | 57.8869768162 |
| md | 1.8694559308 | 0.0000119430 |
| p | 0.7821182923 | 0.0732947275 |
| sigmap1 | 0.0057185724 | 0.0000806263 |
| xi | -43.5851155564 | 3.9426195951 |
| chisq1 | 52.0590011817 | 0.0000000000 |
| chisq2 | 58.5034612697 | 0.0000000000 |
dhad fit faplus -t s -m 203
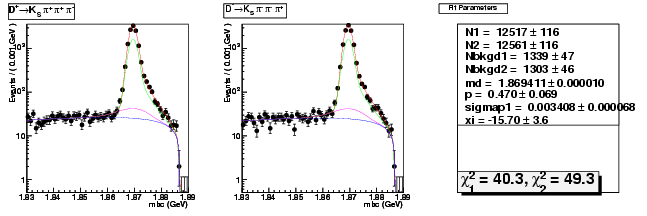
Mode 204 : D+ &rarr KS0 &pi+ &pi+ &pi-
| Name | Value | Error |
|---|---|---|
| N1 | 12517.4834096001 | 115.7586533041 |
| N2 | 12561.4654973248 | 115.8233391124 |
| Nbkgd1 | 1338.7448344921 | 47.1310291945 |
| Nbkgd2 | 1302.7842257870 | 46.4375766071 |
| md | 1.8694105781 | 0.0000102171 |
| p | 0.4697667958 | 0.0693439321 |
| sigmap1 | 0.0034083686 | 0.0000677310 |
| xi | -15.6990806898 | 3.6133594452 |
| chisq1 | 40.3395227119 | 0.0000000000 |
| chisq2 | 49.3443478492 | 0.0000000000 |
dhad fit faplus -t s -m 204
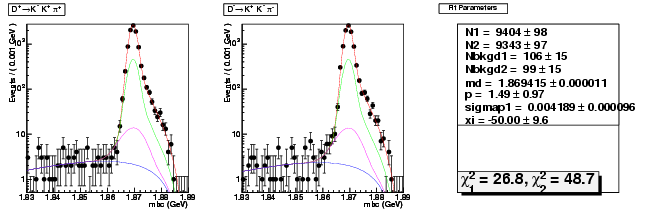
Mode 205 : D+ &rarr K+ K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 9403.9204355387 | 97.6323406897 |
| N2 | 9342.5413125373 | 97.3146658630 |
| Nbkgd1 | 106.1644453656 | 15.3036957639 |
| Nbkgd2 | 99.4967453295 | 15.0691705381 |
| md | 1.8694147088 | 0.0000113173 |
| p | 1.4938903806 | 0.9663883070 |
| sigmap1 | 0.0041888619 | 0.0000958862 |
| xi | -49.9959158115 | 9.5842348020 |
| chisq1 | 26.7828141694 | 0.0000000000 |
| chisq2 | 48.6853988649 | 0.0000000000 |
dhad fit faplus -t s -m 205