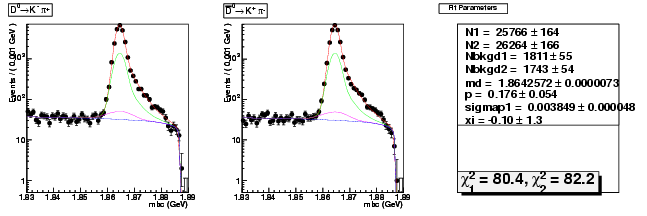
Mode 0 : D0 &rarr K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 25765.6407124416 | 164.1968959013 |
| N2 | 26263.8643613051 | 165.6665824039 |
| Nbkgd1 | 1810.9559313226 | 54.8285182507 |
| Nbkgd2 | 1742.7048044897 | 54.0812198750 |
| md | 1.8642571831 | 0.0000073386 |
| p | 0.1760869963 | 0.0535567681 |
| sigmap1 | 0.0038485158 | 0.0000480313 |
| xi | -0.1000072082 | 1.3306198846 |
| chisq1 | 80.4093141046 | 0.0000000000 |
| chisq2 | 82.2124619576 | 0.0000000000 |
dhad fit saplus -t d -m 0
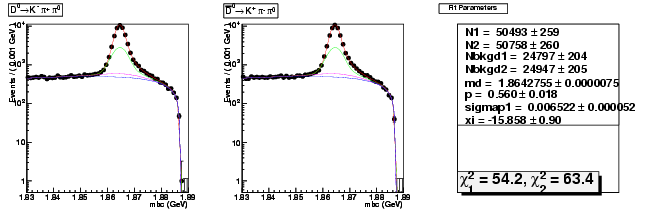
Mode 1 : D0 &rarr K- &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 50492.6738895484 | 259.1376578910 |
| N2 | 50758.2978044250 | 260.4657405941 |
| Nbkgd1 | 24797.4943460482 | 203.6121806942 |
| Nbkgd2 | 24946.8775083828 | 205.0197166367 |
| md | 1.8642755038 | 0.0000074893 |
| p | 0.5602119310 | 0.0179836022 |
| sigmap1 | 0.0065215283 | 0.0000520480 |
| xi | -15.8578730189 | 0.8953784918 |
| chisq1 | 54.1577028960 | 0.0000000000 |
| chisq2 | 63.3884375275 | 0.0000000000 |
dhad fit saplus -t d -m 1
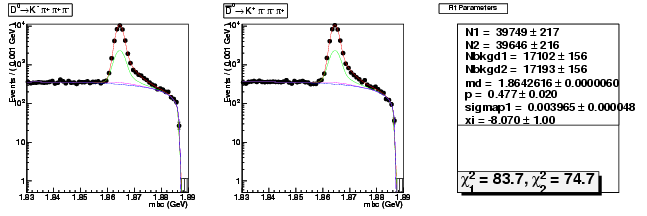
Mode 3 : D0 &rarr K- &pi+ &pi- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 39748.9097458087 | 216.6212330811 |
| N2 | 39646.3881920452 | 216.3433401932 |
| Nbkgd1 | 17102.0213808536 | 155.8148937407 |
| Nbkgd2 | 17192.5489461166 | 156.0476123603 |
| md | 1.8642615527 | 0.0000060059 |
| p | 0.4771102302 | 0.0203666124 |
| sigmap1 | 0.0039649125 | 0.0000479852 |
| xi | -8.0701857911 | 0.9963778060 |
| chisq1 | 83.7266820985 | 0.0000000000 |
| chisq2 | 74.7007561612 | 0.0000000000 |
dhad fit saplus -t d -m 3
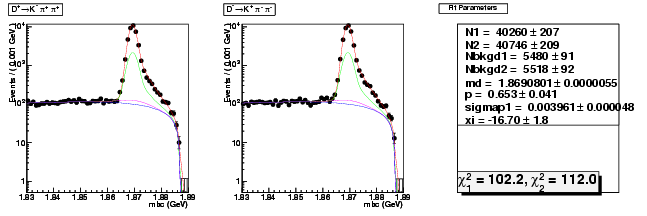
Mode 200 : D+ &rarr K- &pi+ &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 40260.3363392246 | 207.4986262829 |
| N2 | 40745.9537813934 | 208.8032756821 |
| Nbkgd1 | 5479.6353994964 | 90.9788235155 |
| Nbkgd2 | 5518.0172780526 | 91.5045397889 |
| md | 1.8690801347 | 0.0000055380 |
| p | 0.6526288605 | 0.0405335432 |
| sigmap1 | 0.0039606148 | 0.0000480821 |
| xi | -16.7040154249 | 1.8086715086 |
| chisq1 | 102.1721189845 | 0.0000000000 |
| chisq2 | 111.9665011139 | 0.0000000000 |
dhad fit saplus -t d -m 200
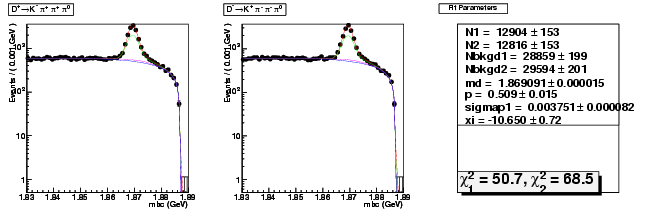
Mode 201 : D+ &rarr K- &pi+ &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 12904.0278244725 | 153.2506632102 |
| N2 | 12815.5485408605 | 153.1728812558 |
| Nbkgd1 | 28858.9734776195 | 198.5447107456 |
| Nbkgd2 | 29594.4586433966 | 200.5482107470 |
| md | 1.8690914778 | 0.0000151828 |
| p | 0.5093599805 | 0.0145573874 |
| sigmap1 | 0.0037514594 | 0.0000824287 |
| xi | -10.6497133608 | 0.7234096365 |
| chisq1 | 50.6674140778 | 0.0000000000 |
| chisq2 | 68.5484259140 | 0.0000000000 |
dhad fit saplus -t d -m 201
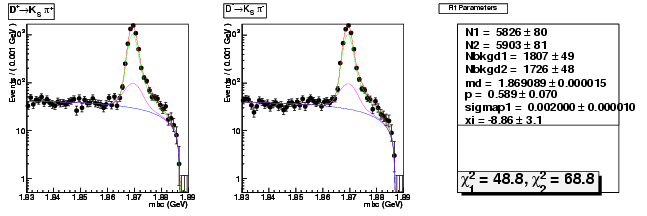
Mode 202 : D+ &rarr KS0 &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 5826.2859008543 | 80.3447311028 |
| N2 | 5903.4891772196 | 80.5285965219 |
| Nbkgd1 | 1807.0854721626 | 49.3557412357 |
| Nbkgd2 | 1725.7480001490 | 48.0333471269 |
| md | 1.8690891739 | 0.0000149327 |
| p | 0.5887787602 | 0.0701850269 |
| sigmap1 | 0.0020000001 | 0.0000101311 |
| xi | -8.8586155029 | 3.1418461873 |
| chisq1 | 48.7702156312 | 0.0000000000 |
| chisq2 | 68.7527432891 | 0.0000000000 |
dhad fit saplus -t d -m 202
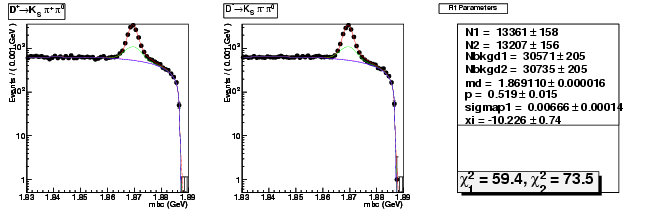
Mode 203 : D+ &rarr KS0 &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 13361.0265487619 | 158.0436910460 |
| N2 | 13206.8777312920 | 156.4431513712 |
| Nbkgd1 | 30571.3356258410 | 205.3908076890 |
| Nbkgd2 | 30735.2436673452 | 204.9382496891 |
| md | 1.8691101528 | 0.0000158725 |
| p | 0.5194513663 | 0.0150322499 |
| sigmap1 | 0.0066619129 | 0.0001442075 |
| xi | -10.2264361410 | 0.7428195361 |
| chisq1 | 59.3679301786 | 0.0000000000 |
| chisq2 | 73.4601521298 | 0.0000000000 |
dhad fit saplus -t d -m 203
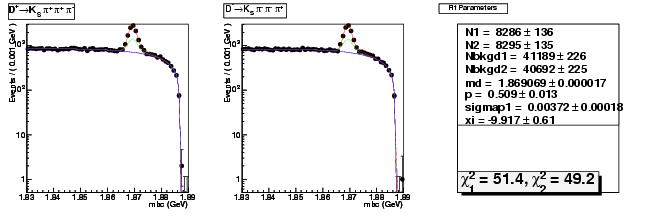
Mode 204 : D+ &rarr KS0 &pi+ &pi+ &pi-
| Name | Value | Error |
|---|---|---|
| N1 | 8285.8913595002 | 135.5728559346 |
| N2 | 8295.3062951042 | 135.1162218346 |
| Nbkgd1 | 41189.2205110497 | 226.4510313340 |
| Nbkgd2 | 40691.8312103698 | 225.0561653372 |
| md | 1.8690694673 | 0.0000174186 |
| p | 0.5086134532 | 0.0125551182 |
| sigmap1 | 0.0037241980 | 0.0001783463 |
| xi | -9.9172111446 | 0.6089763390 |
| chisq1 | 51.4311076387 | 0.0000000000 |
| chisq2 | 49.2062504735 | 0.0000000000 |
dhad fit saplus -t d -m 204
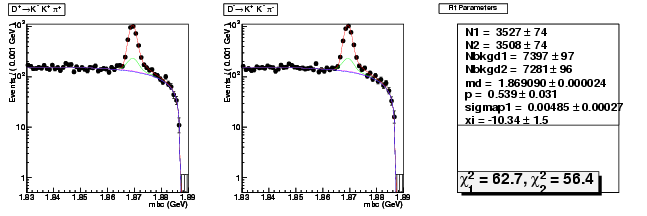
Mode 205 : D+ &rarr K+ K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 3526.8692995258 | 74.2035775778 |
| N2 | 3507.9476795801 | 73.5527271065 |
| Nbkgd1 | 7397.2405834095 | 96.8325273349 |
| Nbkgd2 | 7281.1513967322 | 95.8287029285 |
| md | 1.8690902499 | 0.0000237105 |
| p | 0.5389754365 | 0.0306113768 |
| sigmap1 | 0.0048470999 | 0.0002683120 |
| xi | -10.3419738950 | 1.4694341961 |
| chisq1 | 62.7218657491 | 0.0000000000 |
| chisq2 | 56.3960768432 | 0.0000000000 |
dhad fit saplus -t d -m 205