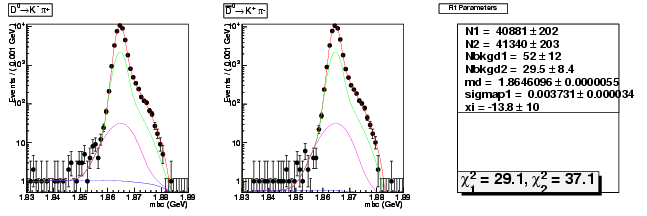
Mode 0 : D0 &rarr K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 40880.7708803462 | 202.4371943193 |
| N2 | 41339.5192876862 | 203.4269164512 |
| Nbkgd1 | 52.2385531923 | 12.3484527429 |
| Nbkgd2 | 29.4877556929 | 8.3567992954 |
| md | 1.8646096490 | 0.0000055414 |
| p | 0.5000000000 | 0.0000000000 |
| sigmap1 | 0.0037309405 | 0.0000339153 |
| xi | -13.7513289470 | 10.0447897277 |
| chisq1 | 29.1345880144 | 0.0000000000 |
| chisq2 | 37.1491571562 | 0.0000000000 |
dhad fit saplus -t s -m 0
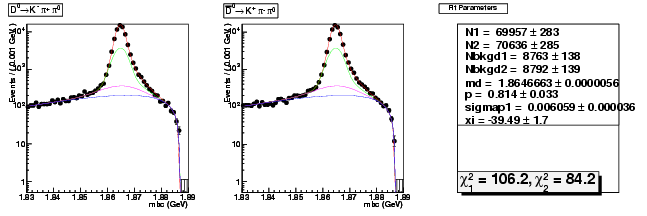
Mode 1 : D0 &rarr K- &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 69957.1584055912 | 283.1550306611 |
| N2 | 70636.4891244121 | 284.6862323731 |
| Nbkgd1 | 8762.5725098795 | 137.7928415009 |
| Nbkgd2 | 8791.9169918543 | 138.5796201022 |
| md | 1.8646663090 | 0.0000055787 |
| p | 0.8137926692 | 0.0330606478 |
| sigmap1 | 0.0060592594 | 0.0000361626 |
| xi | -39.4897212890 | 1.6891949686 |
| chisq1 | 106.2303459831 | 0.0000000000 |
| chisq2 | 84.1764714360 | 0.0000000000 |
dhad fit saplus -t s -m 1
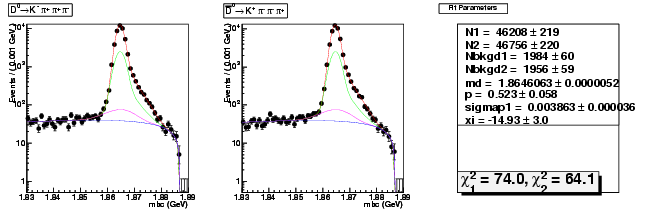
Mode 3 : D0 &rarr K- &pi+ &pi- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 46207.7314676509 | 218.6518162763 |
| N2 | 46756.1011595077 | 219.8464746116 |
| Nbkgd1 | 1983.9693683422 | 59.9107388880 |
| Nbkgd2 | 1955.6057081285 | 59.4670583323 |
| md | 1.8646063428 | 0.0000052153 |
| p | 0.5227060442 | 0.0578403092 |
| sigmap1 | 0.0038628672 | 0.0000362289 |
| xi | -14.9254472038 | 3.0189435144 |
| chisq1 | 74.0357454199 | 0.0000000000 |
| chisq2 | 64.1029478281 | 0.0000000000 |
dhad fit saplus -t s -m 3
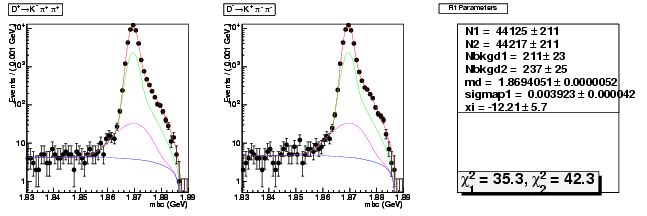
Mode 200 : D+ &rarr K- &pi+ &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 44125.3192829724 | 210.8380142501 |
| N2 | 44217.3132117253 | 211.1486748877 |
| Nbkgd1 | 211.4558678805 | 23.1937102433 |
| Nbkgd2 | 237.4243979658 | 24.5813721208 |
| md | 1.8694050711 | 0.0000051565 |
| p | 0.5000000000 | 0.0000000000 |
| sigmap1 | 0.0039231544 | 0.0000417135 |
| xi | -12.2108573894 | 5.6631581926 |
| chisq1 | 35.2999502090 | 0.0000000000 |
| chisq2 | 42.2721571077 | 0.0000000000 |
dhad fit saplus -t s -m 200
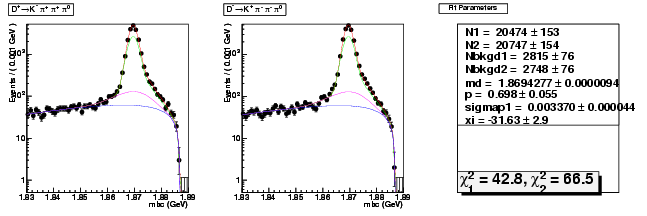
Mode 201 : D+ &rarr K- &pi+ &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 20473.9252303671 | 152.9166275742 |
| N2 | 20746.8177952384 | 153.9856937727 |
| Nbkgd1 | 2815.2328621176 | 75.6652292999 |
| Nbkgd2 | 2748.3687792256 | 75.5855104118 |
| md | 1.8694276796 | 0.0000093668 |
| p | 0.6981113416 | 0.0549286609 |
| sigmap1 | 0.0033704986 | 0.0000436859 |
| xi | -31.6252100444 | 2.8646679120 |
| chisq1 | 42.7771388823 | 0.0000000000 |
| chisq2 | 66.4572950144 | 0.0000000000 |
dhad fit saplus -t s -m 201
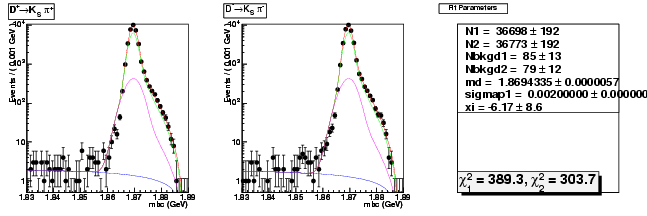
Mode 202 : D+ &rarr KS0 &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 36698.2766331815 | 191.7729509983 |
| N2 | 36773.2049339955 | 191.9641508055 |
| Nbkgd1 | 84.7329016063 | 12.7835714549 |
| Nbkgd2 | 78.7821349566 | 12.4775805771 |
| md | 1.8694335314 | 0.0000057060 |
| p | 0.5000000000 | 0.0000000000 |
| sigmap1 | 0.0020000000 | 0.0000003880 |
| xi | -6.1732257365 | 8.6338984911 |
| chisq1 | 389.2643494338 | 0.0000000000 |
| chisq2 | 303.6785266807 | 0.0000000000 |
dhad fit saplus -t s -m 202
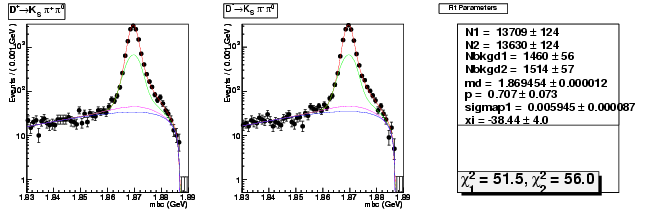
Mode 203 : D+ &rarr KS0 &pi+ &pi0
| Name | Value | Error |
|---|---|---|
| N1 | 13709.3691446283 | 124.2073701512 |
| N2 | 13630.0769335804 | 124.0777378272 |
| Nbkgd1 | 1459.8442306080 | 56.3720599384 |
| Nbkgd2 | 1514.0224532099 | 57.2660839905 |
| md | 1.8694543570 | 0.0000120888 |
| p | 0.7067310263 | 0.0734990491 |
| sigmap1 | 0.0059448148 | 0.0000871089 |
| xi | -38.4360609078 | 3.9966690234 |
| chisq1 | 51.5199134207 | 0.0000000000 |
| chisq2 | 56.0185047710 | 0.0000000000 |
dhad fit saplus -t s -m 203
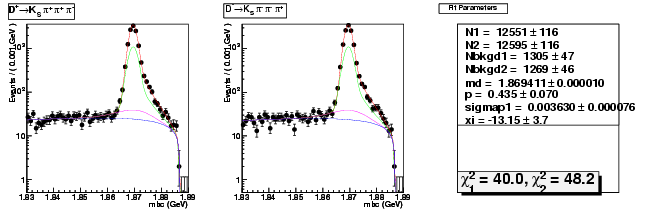
Mode 204 : D+ &rarr KS0 &pi+ &pi+ &pi-
| Name | Value | Error |
|---|---|---|
| N1 | 12551.4155631202 | 116.0273546471 |
| N2 | 12595.4401207535 | 116.0825970112 |
| Nbkgd1 | 1304.6150864925 | 47.0684054996 |
| Nbkgd2 | 1268.6108825780 | 46.3490983150 |
| md | 1.8694106347 | 0.0000101577 |
| p | 0.4351159578 | 0.0698126367 |
| sigmap1 | 0.0036304245 | 0.0000762642 |
| xi | -13.1507632999 | 3.6602976557 |
| chisq1 | 39.9749908261 | 0.0000000000 |
| chisq2 | 48.1565207562 | 0.0000000000 |
dhad fit saplus -t s -m 204
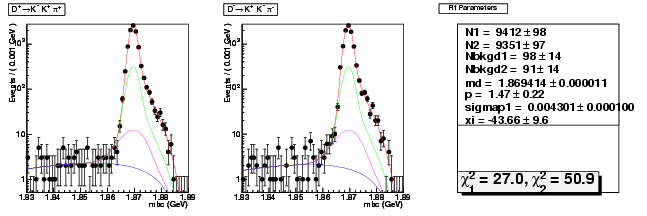
Mode 205 : D+ &rarr K+ K- &pi+
| Name | Value | Error |
|---|---|---|
| N1 | 9411.7794706169 | 97.5744725668 |
| N2 | 9351.2519108922 | 97.2642514368 |
| Nbkgd1 | 98.4607866022 | 14.3998788391 |
| Nbkgd2 | 90.9013740834 | 14.1383119903 |
| md | 1.8694143047 | 0.0000112133 |
| p | 1.4712589648 | 0.2201843927 |
| sigmap1 | 0.0043011646 | 0.0000999983 |
| xi | -43.6615803295 | 9.6061382544 |
| chisq1 | 27.0151183849 | 0.0000000000 |
| chisq2 | 50.9370947022 | 0.0000000000 |
dhad fit saplus -t s -m 205