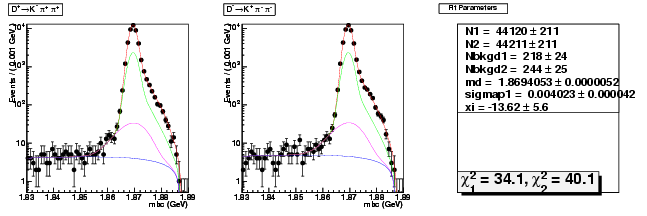
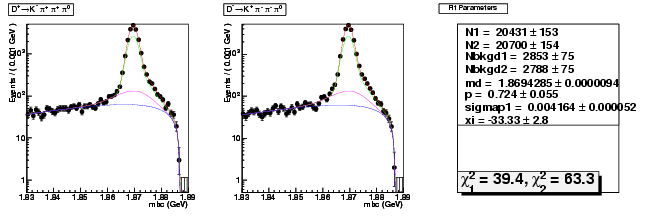
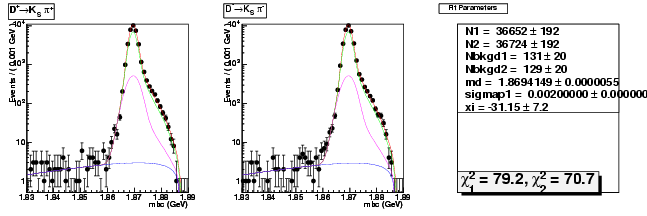
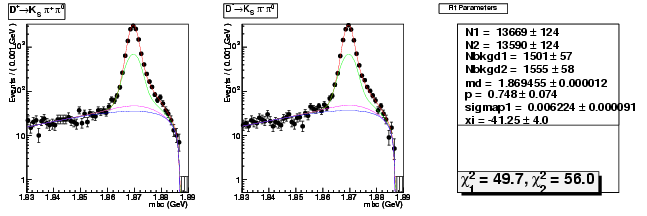
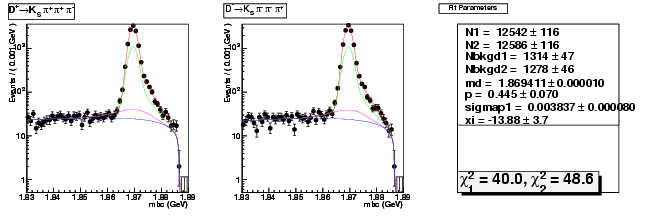
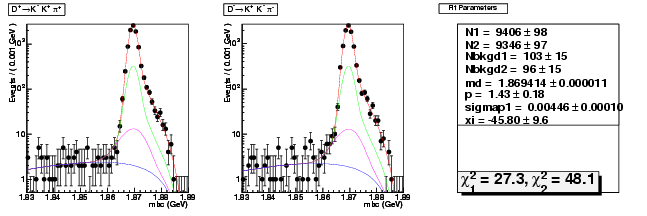
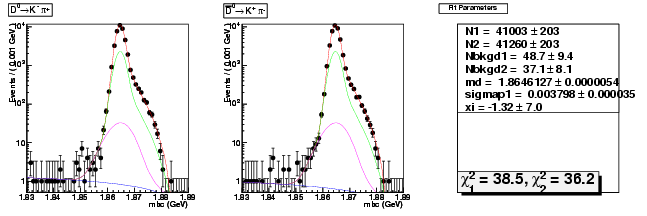
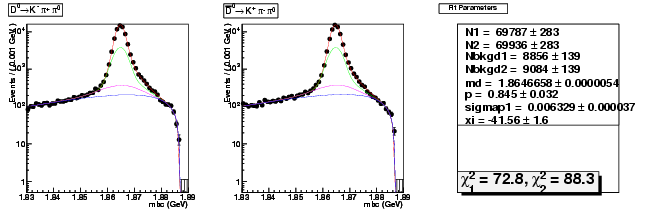
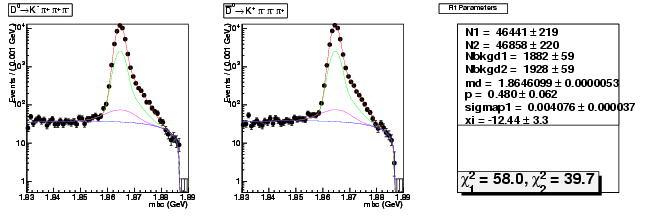
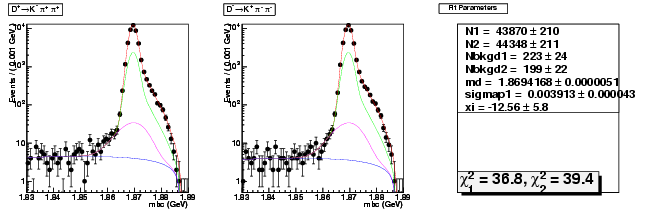
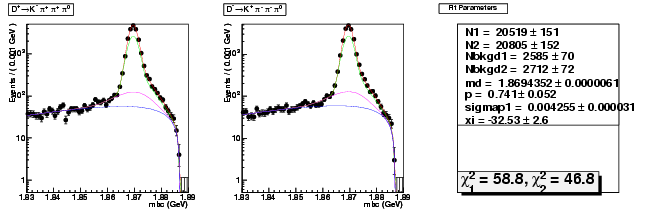
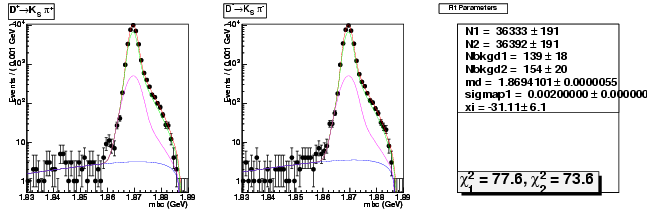
D-Hadronic Analysis - 7.06
Table of Contents
- Overview
- Plots
- Tables
- Data 818/pb
- Data 537/pb
- Compare the 537ipb yield with 281ipb 537ipb in Data
- Compare parameter "sigmap1" data 281ipb 537ipb
- Compare parameter "md" data 281ipb 537ipb
- Compare parameter "p" data 281ipb 537ipb
- Compare parameter "xi" data 281ipb 537ipb
- Compare parameter "chisq1" data 281ipb 537ipb
- Compare parameter "chisq2" data 281ipb 537ipb
- Details
- Software
- Datasets
- DHad-281 CBX
- Start up
- Generate Signal MC (2.0)
- Generate Signal MC with PHOTOS-2.15 (2.1)
- First Try
- Produce Latest Photos/Dalitz MC
- Find the location of the right code
- Run on the code
- Fix the bad pass2 runs
cleog_Single_D0B_to_Kpipipi_6.pdscleog_Single_D0_to_Kpipi0_8.pds- Fix the rest
- Run Dtuple
- Compare yields with photos 2.15
- Check the source of three modes
- Add error to the difference
- Add more digits to the table
- Generate double tag signal MC (2.1) [%]
- Process 281 data (2.1)
- Process 818 data (2.2) [88%]
- First Try on data 43,44,45,46
- Set up src 2.2
- Second Try : Encounter the coredump
- Trace the coredump: CPU time limit exceeded
- Trace the "To many missing masses" problem
- Break down data43 into two
- Event Display the trouble event
- Inplement code to bypass the trouble events
- Avoid missing mass calc
- Use Skip Bad Event
- Proceed on the rest of the data43
- Avoid the memeory leak for data43
- Put Time Statement inside code
- Proceed on data44, 45, 46
- Process data44
- Process data46
- Re-process data43
- Re-process data45
- Extract Yields
- Do the Fits
- Store the fitting plots
- Extract yields for 818 data
- Fitting for 818 data
- Store fitting plots
- Compare the yields
- Compare the fitting parameters
- Process 281 data with eventstore 20070822 (2.2)
- Prepare four-vector for CLEOG (2.3)
- Debug in the BeamEnergyProxy (2.4)
- Fit 537 data with new parameters (2.5)
Overview
Tables
Data 818/pb
| Mode | data yield (281ipb) | data yield (818ipb) | 818ipb/281ipb |
|---|---|---|---|
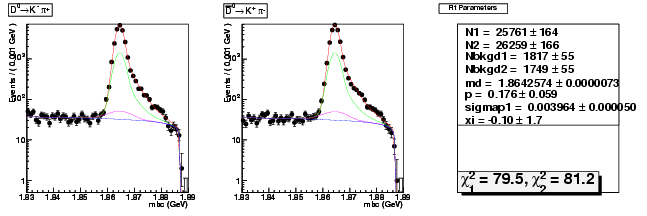
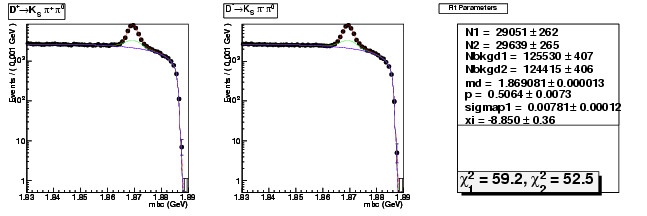
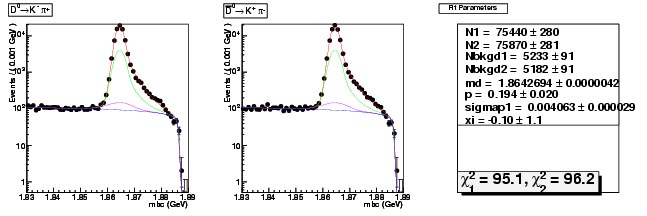
| D0→ K-π+ | 25761 ± 164 | 75440 ± 280 | 2.93 ± 0.02 |
| D0B→ K+ π- | 26259 ± 166 | 75870 ± 281 | 2.89 ± 0.02 |
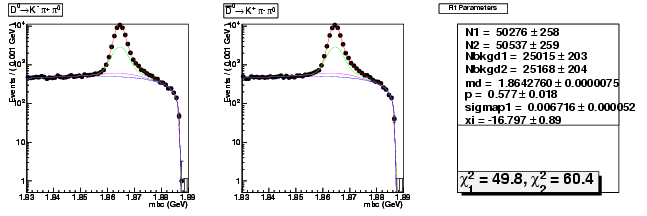
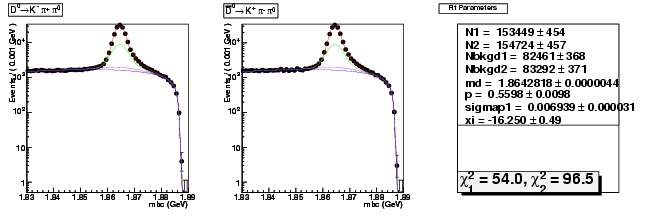
| D0→ K-π+ π0 | 50276 ± 258 | 153449 ± 454 | 3.05 ± 0.02 |
| D0B→ K+ π-π0 | 50537 ± 259 | 154724 ± 457 | 3.06 ± 0.02 |
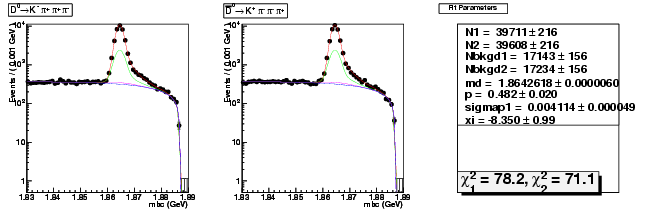
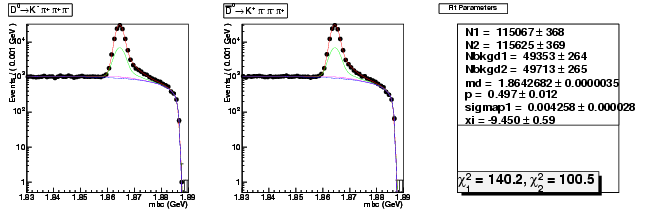
| D0→ Kπ+ ππ+ | 39711 ± 216 | 115067 ± 368 | 2.90 ± 0.02 |
| D0B→ K+ ππ+ π | 39608 ± 216 | 115625 ± 369 | 2.92 ± 0.02 |
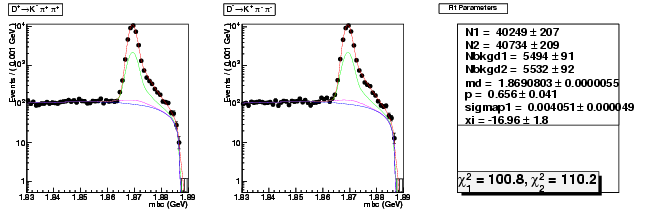
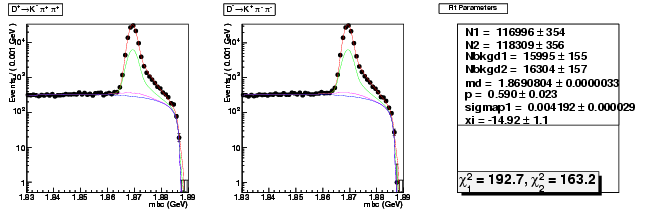
| D+ → K-π+ π+ | 40249 ± 207 | 116996 ± 354 | 2.91 ± 0.02 |
| D→ K+ ππ- | 40734 ± 209 | 118309 ± 356 | 2.90 ± 0.02 |
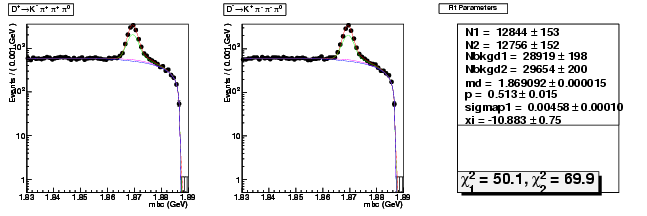
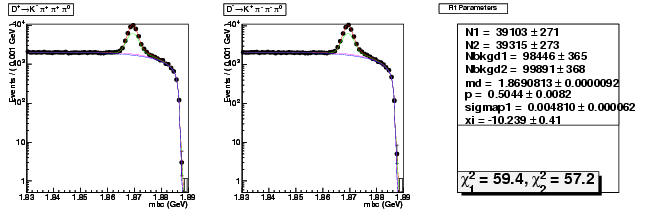
| D+ → K-π+ π+ π0 | 12844 ± 153 | 39103 ± 271 | 3.04 ± 0.04 |
| D→ K+ ππ-π0 | 12756 ± 152 | 39315 ± 273 | 3.08 ± 0.04 |
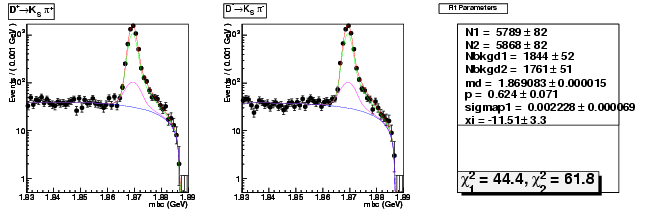
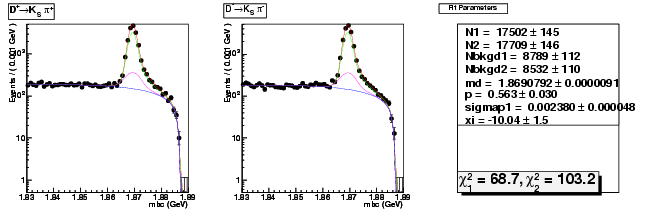
| D+ → K0Sπ+ | 5789 ± 82 | 17502 ± 145 | 3.02 ± 0.05 |
| D→ K0Sπ | 5868 ± 82 | 17709 ± 146 | 3.02 ± 0.05 |
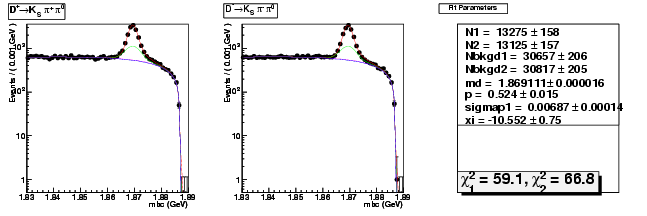
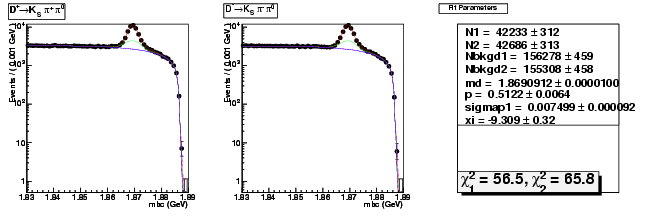
| D+ → K0Sπ+ π0 | 13275 ± 158 | 42233 ± 312 | 3.18 ± 0.04 |
| D→ K0Sππ0 | 13125 ± 157 | 42686 ± 313 | 3.25 ± 0.05 |
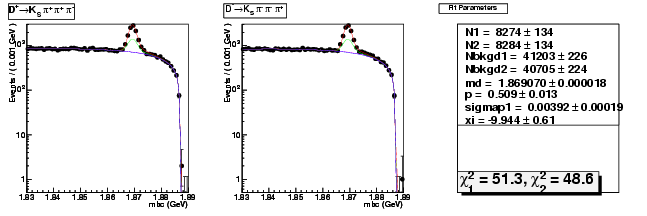
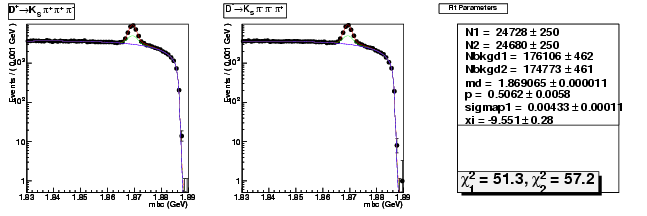
| D+ → K0Sπ+ π+ π- | 8274 ± 134 | 24728 ± 250 | 2.99 ± 0.06 |
| D→ K0Sππ-π+ | 8284 ± 134 | 24680 ± 250 | 2.98 ± 0.06 |
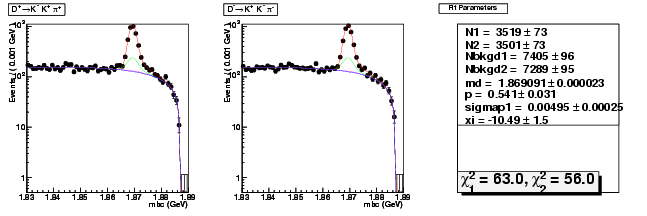
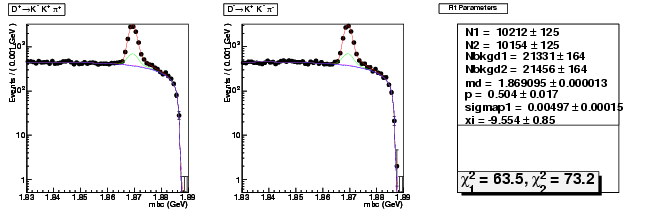
| D+ → K+ K-π+ | 3519 ± 73 | 10212 ± 125 | 2.90 ± 0.07 |
| D→ KK+π- | 3501 ± 73 | 10154 ± 125 | 2.90 ± 0.07 |
818/281 = 2.91
dhad-2.2 table compare yields data divide 281ipb 818ipb
Data 537/pb
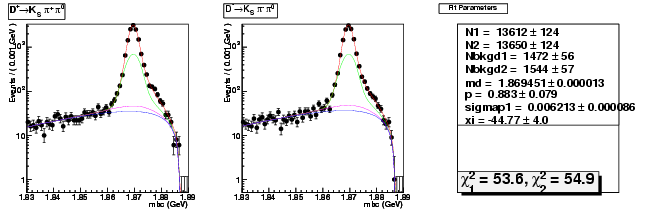
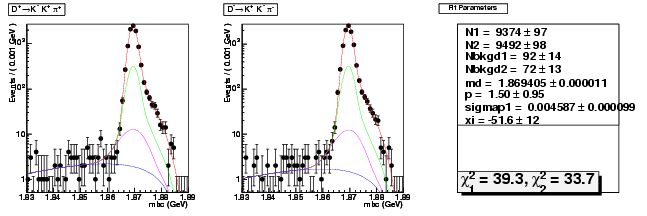
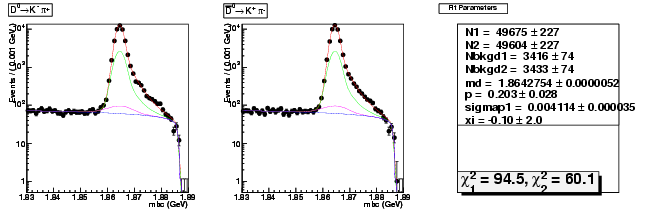
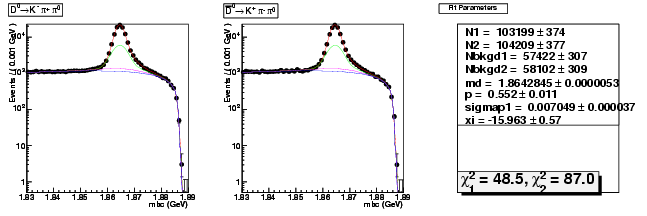
Compare the 537ipb yield with 281ipb 537ipb in Data
| Mode | data yield (281ipb) | data yield (537ipb) | 537ipb/281ipb |
|---|---|---|---|
| D0→ K-π+ | 25761 ± 164 | 49675 ± 227 | 1.93 ± 0.02 |
| D0B→ K+ π- | 26259 ± 166 | 49604 ± 227 | 1.89 ± 0.01 |
| D0→ K-π+ π0 | 50276 ± 258 | 103199 ± 374 | 2.05 ± 0.01 |
| D0B→ K+ π-π0 | 50537 ± 259 | 104209 ± 377 | 2.06 ± 0.01 |
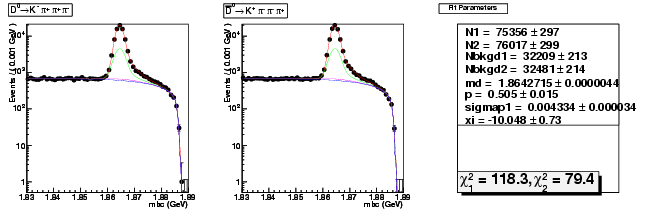
| D0→ Kπ+ ππ+ | 39711 ± 216 | 75356 ± 297 | 1.90 ± 0.01 |
| D0B→ K+ ππ+ π | 39608 ± 216 | 76017 ± 299 | 1.92 ± 0.01 |
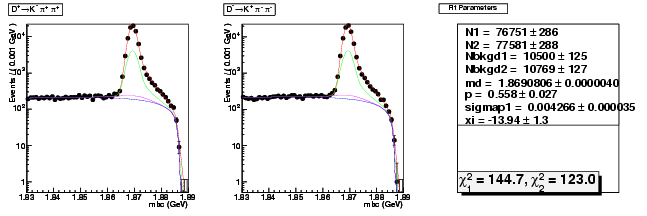
| D+ → K-π+ π+ | 40249 ± 207 | 76751 ± 286 | 1.91 ± 0.01 |
| D→ K+ ππ- | 40734 ± 209 | 77581 ± 288 | 1.90 ± 0.01 |
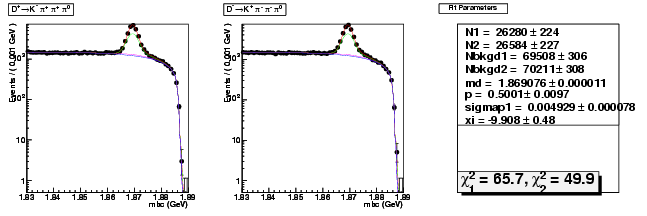
| D+ → K-π+ π+ π0 | 12844 ± 153 | 26280 ± 224 | 2.05 ± 0.03 |
| D→ K+ ππ-π0 | 12756 ± 152 | 26584 ± 227 | 2.08 ± 0.03 |
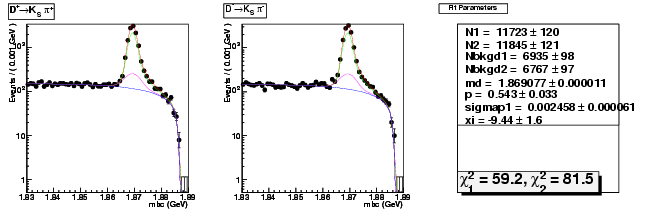
| D+ → K0Sπ+ | 5789 ± 82 | 11723 ± 120 | 2.03 ± 0.04 |
| D→ K0Sπ | 5868 ± 82 | 11845 ± 121 | 2.02 ± 0.03 |
| D+ → K0Sπ+ π0 | 13275 ± 158 | 29051 ± 262 | 2.19 ± 0.03 |
| D→ K0Sππ0 | 13125 ± 157 | 29639 ± 265 | 2.26 ± 0.03 |
| D+ → K0Sπ+ π+ π- | 8274 ± 134 | 16506 ± 214 | 1.99 ± 0.04 |
| D→ K0Sππ-π+ | 8284 ± 134 | 16449 ± 215 | 1.99 ± 0.04 |
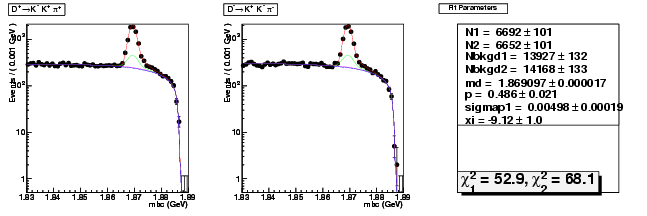
| D+ → K+ K-π+ | 3519 ± 73 | 6692 ± 101 | 1.90 ± 0.05 |
| D→ KK+π- | 3501 ± 73 | 6652 ± 101 | 1.90 ± 0.05 |
537/281 = 1.91
copied from: dhad-2.2 table compare yields data divide 281ipb 537ipb
537/281 = 1.91
dhad-2.2 table compare yields data divide 281ipb 537ipb
Compare parameter "sigmap1" data 281ipb 537ipb
| Mode | 281ipb | 537ipb | diff(%) |
|---|---|---|---|
| D0→ K-π+ | 0.003964 ± 0.000050 | 0.004114 ± 0.000035 | +3.8(+3.0σ) |
| D0→ K-π+ π0 | 0.006716 ± 0.000052 | 0.007049 ± 0.000037 | +5.0(+6.4σ) |
| D0→ Kπ+ ππ+ | 0.004114 ± 0.000049 | 0.004334 ± 0.000034 | +5.3(+4.5σ) |
| D+ → K-π+ π+ | 0.004051 ± 0.000049 | 0.004266 ± 0.000035 | +5.3(+4.4σ) |
| D+ → K-π+ π+ π0 | 0.004579 ± 0.000102 | 0.004929 ± 0.000078 | +7.6(+3.4σ) |
| D+ → K0Sπ+ | 0.002228 ± 0.000069 | 0.002458 ± 0.000061 | +10.3(+3.3σ) |
| D+ → K0Sπ+ π0 | 0.006869 ± 0.000144 | 0.007814 ± 0.000122 | +13.8(+6.6σ) |
| D+ → K0Sπ+ π+ π- | 0.003918 ± 0.000187 | 0.004565 ± 0.000143 | +16.5(+3.5σ) |
| D+ → K+ K-π+ | 0.004948 ± 0.000248 | 0.004978 ± 0.000192 | +0.6(+0.1σ) |
dhad-2.2 table compare parameter sigmap1 data 281ipb 537ipb --set rnd=0.000001
Compare parameter "md" data 281ipb 537ipb
| Mode | 281ipb | 537ipb | diff(%) |
|---|---|---|---|
| D0→ K-π+ | 1.864257 ± 0.000007 | 1.864275 ± 0.000005 | 0.0(+2.6σ) |
| D0→ K-π+ π0 | 1.864276 ± 0.000008 | 1.864284 ± 0.000005 | 0.0(+1.0σ) |
| D0→ Kπ+ ππ+ | 1.864262 ± 0.000006 | 1.864272 ± 0.000004 | 0.0(+1.7σ) |
| D+ → K-π+ π+ | 1.869080 ± 0.000006 | 1.869081 ± 0.000004 | 0.0(+0.2σ) |
| D+ → K-π+ π+ π0 | 1.869092 ± 0.000015 | 1.869076 ± 0.000011 | -0.0(-1.1σ) |
| D+ → K0Sπ+ | 1.869083 ± 0.000015 | 1.869077 ± 0.000011 | -0.0(-0.4σ) |
| D+ → K0Sπ+ π0 | 1.869111 ± 0.000016 | 1.869081 ± 0.000013 | -0.0(-1.9σ) |
| D+ → K0Sπ+ π+ π- | 1.869070 ± 0.000018 | 1.869062 ± 0.000015 | -0.0(-0.4σ) |
| D+ → K+ K-π+ | 1.869091 ± 0.000023 | 1.869097 ± 0.000017 | 0.0(+0.3σ) |
dhad-2.2 table compare parameter md data 281ipb 537ipb --set rnd=0.000001
Compare parameter "p" data 281ipb 537ipb
| Mode | 281ipb | 537ipb | diff(%) |
|---|---|---|---|
| D0→ K-π+ | 0.176 ± 0.059 | 0.203 ± 0.028 | +15.3(+0.5σ) |
| D0→ K-π+ π0 | 0.577 ± 0.018 | 0.552 ± 0.011 | -4.3(-1.4σ) |
| D0→ Kπ+ ππ+ | 0.482 ± 0.020 | 0.505 ± 0.015 | +4.8(+1.2σ) |
| D+ → K-π+ π+ | 0.656 ± 0.041 | 0.558 ± 0.027 | -14.9(-2.4σ) |
| D+ → K-π+ π+ π0 | 0.513 ± 0.015 | 0.500 ± 0.010 | -2.5(-0.9σ) |
| D+ → K0Sπ+ | 0.624 ± 0.071 | 0.543 ± 0.033 | -13.0(-1.1σ) |
| D+ → K0Sπ+ π0 | 0.524 ± 0.015 | 0.506 ± 0.007 | -3.4(-1.2σ) |
| D+ → K0Sπ+ π+ π- | 0.509 ± 0.013 | 0.504 ± 0.007 | -1.0(-0.4σ) |
| D+ → K+ K-π+ | 0.541 ± 0.031 | 0.486 ± 0.021 | -10.2(-1.8σ) |
dhad-2.2 table compare parameter p data 281ipb 537ipb --set rnd=0.001
Compare parameter "xi" data 281ipb 537ipb
| Mode | 281ipb | 537ipb | diff(%) |
|---|---|---|---|
| D0→ K-π+ | -0.100 ± 1.655 | -0.100 ± 1.981 | -0.0(0.0σ) |
| D0→ K-π+ π0 | -16.797 ± 0.891 | -15.963 ± 0.572 | -5.0(+0.9σ) |
| D0→ Kπ+ ππ+ | -8.350 ± 0.994 | -10.048 ± 0.733 | +20.3(-1.7σ) |
| D+ → K-π+ π+ | -16.961 ± 1.808 | -13.944 ± 1.255 | -17.8(+1.7σ) |
| D+ → K-π+ π+ π0 | -10.883 ± 0.746 | -9.908 ± 0.483 | -9.0(+1.3σ) |
| D+ → K0Sπ+ | -11.510 ± 3.290 | -9.438 ± 1.622 | -18.0(+0.6σ) |
| D+ → K0Sπ+ π0 | -10.552 ± 0.751 | -8.850 ± 0.362 | -16.1(+2.3σ) |
| D+ → K0Sπ+ π+ π- | -9.944 ± 0.605 | -9.348 ± 0.325 | -6.0(+1.0σ) |
| D+ → K+ K-π+ | -10.487 ± 1.459 | -9.123 ± 1.041 | -13.0(+0.9σ) |
dhad-2.2 table compare parameter xi data 281ipb 537ipb --set rnd=0.001
Compare parameter "chisq1" data 281ipb 537ipb
| Mode | 281ipb | 537ipb | diff(%) |
|---|---|---|---|
| D0→ K-π+ | 79.473 ± 0.000 | 94.482 ± 0.000 | 18.9 |
| D0→ K-π+ π0 | 49.782 ± 0.000 | 48.453 ± 0.000 | -2.7 |
| D0→ Kπ+ ππ+ | 78.229 ± 0.000 | 118.298 ± 0.000 | 51.2 |
| D+ → K-π+ π+ | 100.756 ± 0.000 | 144.695 ± 0.000 | 43.6 |
| D+ → K-π+ π+ π0 | 50.059 ± 0.000 | 65.742 ± 0.000 | 31.3 |
| D+ → K0Sπ+ | 44.374 ± 0.000 | 59.182 ± 0.000 | 33.4 |
| D+ → K0Sπ+ π0 | 59.143 ± 0.000 | 59.199 ± 0.000 | 0.1 |
| D+ → K0Sπ+ π+ π- | 51.269 ± 0.000 | 53.187 ± 0.000 | 3.7 |
| D+ → K+ K-π+ | 63.001 ± 0.000 | 52.884 ± 0.000 | -16.1 |
dhad-2.2 table compare parameter chisq1 data 281ipb 537ipb --set rnd=0.001
Compare parameter "chisq2" data 281ipb 537ipb
| Mode | 281ipb | 537ipb | diff(%) |
|---|---|---|---|
| D0B→ K+ π- | 81.193 ± 0.000 | 60.059 ± 0.000 | -26.0 |
| D0B→ K+ π-π0 | 60.430 ± 0.000 | 87.035 ± 0.000 | 44.0 |
| D0B→ K+ ππ+ π | 71.091 ± 0.000 | 79.385 ± 0.000 | 11.7 |
| D→ K+ ππ- | 110.211 ± 0.000 | 122.951 ± 0.000 | 11.6 |
| D→ K+ ππ-π0 | 69.903 ± 0.000 | 49.937 ± 0.000 | -28.6 |
| D→ K0Sπ | 61.790 ± 0.000 | 81.452 ± 0.000 | 31.8 |
| D→ K0Sππ0 | 66.779 ± 0.000 | 52.488 ± 0.000 | -21.4 |
| D→ K0Sππ-π+ | 48.635 ± 0.000 | 69.994 ± 0.000 | 43.9 |
| D→ KK+π- | 55.973 ± 0.000 | 68.069 ± 0.000 | 21.6 |
dhad-2.2 table compare parameter chisq2 data 281ipb 537ipb --set rnd=0.001,namestyle=fnamebar
Details
Software
-
CLEOG :
20050525_MCGEN -
PASS2 :
20041104_MCP2 -
NTuple :
20050316_FULL
Datasets
281/pb :
| datasets | lumi (/pb) |
|---|---|
| data31 | 21.5 |
| data32 | 32.4 |
| data33 | 6.5 |
| data35 | 52.0 |
| data36 | 70.7 |
| data37 | 111.1 |
Extened (818/pb):
| datasets | lumi (/pb) |
|---|---|
| data43 | 125.154 |
| data44 | 187.754 |
| data45 | 118.153 |
| data46 | 148.579 |
DHad-281 CBX
-
Update CBX
- Read the last document for updating on page.
-
Try:
dhad update cbx tableAdd local package to setup.sh.
Script is working.
-
Need to check the source and result.
-
Table 36: tab:fitResultsData
From the DHadAttr.py found
cbx_data_results_pdgReproduce this table
dhad table cbx_data_results_pdg
Found error:
+-0.0-0.1sigmaFix it … done.
Update to K0S, Reference:
table prd_data_results K0Sdhad table cbx_data_results_pdg K0S
Done.
-
Table 37: tab:fitResultsRatiosData
cbx_data_results_bf_ratio_pdgdhad table cbx_data_results_bf_ratio_pdg K0S
Done.
-
Table 38: tab:correlationMatrixData
dhad table cbx_data_correlation_matrix K0S
Done.
-
Table 39: tab:yieldSTResidualsData
dhad table cbx_residual_single K0S
Done.
-
Table 40: tab:yieldDTResidualsData
dhad table cbx_residual_double K0S
Done.
-
Table 41: tab:fitResultsDataVariations
dhad table cbx_data_results_variations K0S
Done.
-
Table 36: tab:fitResultsData
-
Update the CBX tables.
Edit the tables in the DHadAttr.py
dhad update cbx table
Done.
PS file is OK.
Translate all the eps figure to pdf.
dhad update cbx epstopdf ... OK.
PDF file is OK.
-
Check in the latex file and figures
Done. Email to Anders and David. OK.
-
Release the document
From page https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/DocumentDatabase
CBX2009-056
Edit the CBX.
Submitted. In the link:
https://redms.classe.cornell.edu/record/1965
Edit the DHad page … OK.
Edit the Twiki page … done.
Start up
List the Software status
-
What I have
- Yield extraction from DHad NTuple
- Fits for tag yields
- Branching Fraction fitter
- Scripts creating most of the tables for CBX and PRD
-
What I don't have (know)
- How to run the DHad code to produce the DHad NTuple
- How to produce the root squre scaled plots in ROOT
Retrieve Source code
-
Ask Peter … sent.
Reply from Peter:
The directories in question were archived as per ticket #16977. They are about a terabyte worth of (primarily) MC datasets and data/MC ntuples. I don't think much code was involved (possibly some of my stuff for estimating backgrounds etc, but that should be redone for the full dataset anyway).
-
Ask DLK … sent.
Reply from David:
Bill Brangan is the keeper of backups. Easiest way to reach him is to send an email request to service-lepp. Be sure to specify the file names (or subdirectory) and what directory it was in. I hope this was in someone's personal home disk area so it is readily available on backup. Otherwise you may be in trouble. Are you sure that it has migrated to tape? Was it ever committed to the CLEO cvs repository? dlk
Message from Werner:
Can you say what exactly is missing? Was this on a common tem disk? Private code/data/etc in a user's area is not usually moved to tape without notification.
Need to explore the source code!
Check out source code
Start from Software Doc page
- https://www.lepp.cornell.edu/restricted/CLEO/CLEO3/soft/intro/Tutorial/index.html
-
Peter's talk http://www.lns.cornell.edu/public/CLEO/CLEO101/2007/Day5a/070614_cleo101.pdf
Day 1. Richard Gray:
Check the current release:
c3rel
Use this
20080228_FULLin the setdhad.shDay 3. Peter's talk:
D0toKpi example.
-
Ask help from Werner
Sent email to Werner.
Reply from Werner:
To get Peter's ntuple-making processor, do: setenv CVSROOT /nfs/cleo3/cvsroot cvs co HadronicDNtupleProc You should be able to compile this like any other CLEO-III code. For the suez job, it looks like you should use HadronicDNtupleProc/Test/loadHadronicDNtupleProc.tcl (see the top of this file for environment variables you need to set), but I would check with Peter to be sure.
Check out the code :
cvs -d /nfs/cleo3/cvsroot co HadronicDNtupleProc
Error message:
cvs checkout: warning: cannot write to history file /nfs/cleo3/cvsroot/CVSROOT/history: Permission denied cvs checkout: Updating HadronicDNtupleProc cvs checkout: failed to create lock directory for `/nfs/cleo3/cvsroot/Offline/src/HadronicDNtupleProc' (/nfs/cleo3/cvsroot/Offline/src/HadronicDNtupleProc/#cvs.lock): Permission denied cvs checkout: failed to obtain dir lock in repository `/nfs/cleo3/cvsroot/Offline/src/HadronicDNtupleProc' cvs [checkout aborted]: read lock failed - giving up
Ask Werner … sent.
Reply:
Try using "cleo3cvs" instead of "cvs".
Check out the code :
export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co HadronicDNtupleProc ... OK.
Need to make the build area … see page.
Re-set the variables in the setup.sh.
c3make=> make.logCheck out the module
CWNFrameworkcleo3cvs co CWNFramework
c3make=> make.log.1Read Peter's webpage: http://www.lepp.cornell.edu/~ponyisi/cwnframework.html
From the MyProc/MyProc directory, run the script CWNFramework/Test/genDefinitions.py . This will autogenerate the header files for the tuple.
From the MyProc directory, run the script CWNFramework/Test/genSkels.py
Still the same compile error.
Ask Peter … sent.
Reply from Peter:
These appear to be template problems with compiling DChain. Are you somehow getting the wrong version of g++?
Try to uncomment the cms environment.
Still the same problem.
Raise question on the CLEO computing HN. sent.
Message from Paras:
I am having a very similar error right now with the same release, which I also can't seem to explain. I just replaced my current piece of code with a backup piece of code that I knew to be working and I have still have the same problem.
Follow up from Paras:
OOPS, I need to compile my code more often... compiling on the proper machine, lnx134, solved the problem for me hopefully that is also Xin's problem?
Messge from Dan:
Are you compiling on lnx134? If not, what do you get on that system? Remember, building CLEO code on SL4+ systems is not supported, and most of our desktops are now SL4+.
Log on to lnx134. Compile OK.
Test Run on data
Get the example tcl file.
suez -f /home/xs32/work/CLEO/analysis/DHad/script/tcl/test.tcl
Error message:
Suez.test> #Load in your very own processor. Suez.test> proc sel HadronicDNtupleProc %% ERROR-DynamicLoader.DLSharedObjectHandler: /home/xs32/tmp/build/20080228_FULL/Linux/shlib/HadronicDNtupleProc.so_20080228_FULL: undefined symbol: _ZNSs4_Rep20_S_empty_rep_storageE %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load HadronicDNtupleProc.
Found another example from Peter's dir:
cp /nfs/cor/user/ponyisi/daf9/simple-dtuple-data31.sh $dhad/script/bash/
Change it to test_data31.sh, can use the loadHadronicDNtupleProc.tcl
suez -f /home/xs32/work/CLEO/analysis/DHad/script/tcl/test.tcl
%% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: can't read "env(INPUTDATA)": no such variable
Use the test_data31.sh …
;Suez.test> proc sel HadronicDNtupleProc %% ERROR-DynamicLoader.DLSharedObjectHandler: /home/xs32/tmp/build/20080228_FULL/Linux/shlib/HadronicDNtupleProc.so_20080228_FULL: undefined symbol: _ZNSs4_Rep20_S_empty_rep_storageE %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load HadronicDNtupleProc.
Use everything …
cd /home/xs32/work/CLEO/analysis/DHad/tmp cp /nfs/cor/user/ponyisi/daf9/2005-3/local_setup .
Edit the localsetup … Notice in the end c3rel 20050316_FULL, use c3rel 20080228_FULL.
%% ERROR-DynamicLoader.DLSharedObjectHandler: /home/xs32/tmp/build/20080228_FULL/Linux/shlib/HadronicDNtupleProc.so_20080228_FULL: undefined symbol: _ZNSs4_Rep20_S_empty_rep_storageE %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load HadronicDNtupleProc.
Ask questions on Dhad HN … sent.
Message from Fan:
You can check your Makefile to make sure that you have already included all necessary Libs in that file.
From Werner:
Do you have the full log file?
From Sourik:
You could use "c++filt" to digest error message.
From Peter:
You're sure you compiled this on lnx134? This is some kind of C++ string problem (as you can see by running c++filt _ZNSs4_Rep20_S_empty_rep_storageE) so it sounds possibly like a compiler incompatibility.
Check the CBX "code" section:
Monte Carlo was generated with the following prescription: [Cleog] was run using release 20050525_MCGEN. [MCPass2] was run using release 20041104_MCP2 The creation of ntuples was done in the 20050316_FULL release, with the following additional package tags: MCCCTagger v02_00_04 HadronicDNtupleProc ponyisi060929
Check out the additional packages :
export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co -rv02_00_04 MCCCTagger cleo3cvs co -rponyisi060929 HadronicDNtupleProc
Compile with 20050316_FULL release at lnx134:
c3rel 20050316_FULL
export USER_BUILD=/home/xs32/tmp/build/$C3LIB
export USER_SHLIB=${USER_BUILD}/Linux/shlib
c3make
Need to check the CWNFramework:
cleo3cvs co CWNFramework
Compile OK.
Test :
. ./test_data31.sh >& $dhad/log/2009/0210/build.txt
Error build.txt:
Compile again, and test, same result.
Ask on HN… sent.
Message from Surik:
Try to use debugger to find the cause of "abort signal." There is well documented CLEO note, which will help you to DEBUG your code. https://www.lepp.cornell.edu/restricted/CLEO/CLEO3/soft/howto/howto_debugSharedProcessor.html Hope this helps.
Sent reply.
Message from Werner:
You're running in 20050316_FULL, but somehow, you linked against the 20080228_FULL version of ROOT: [ Look for /nfs/cleo3/Offline/rel/20080228_FULL/other_sources/Root/lib/libTree.so in your build.txt ] This may not be the cause of the problem, but certainly looks suspicious.
Trace the ROOT lib …
which root /nfs/cleo3/Offline/rel/20080228_FULL/other_sources/Root/bin/root
Comment out the ROOT and Python part in the setup.sh.
Log off the terminal and restart on lnx224: => build.txt
setdhad cd /home/xs32/work/CLEO/analysis/DHad/test/script . ./test_data31.sh >& $dhad/log/2009/0211/build.txt
%% ERROR-DynamicLoader.DLSharedObjectHandler: HadronicDNtupleProc.so_20050316_FULL: cannot open shared object file: No such file or directory %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load HadronicDNtupleProc.
Use c3rel 20050316_FULL for general now. On lnx224: => build.txt.1 OK.
setdhad . /home/xs32/work/CLEO/analysis/DHad/test/script/test_data31.sh >& $dhad/log/2009/0211/build.txt.1
***** Summary Info *****
Stream beginrun : 1 Stream event : 11 Stream startrun : 1 Processed 13 stops.
Try not use the setdhad in lnx224: => build.txt.2 same problem.
Change the sequence of the settings in the test_data31.sh same as
the defult one. => build.txt.3 same problem.
Use all the same as defult one. (i.e. must use the c3rel $C3LIB again!) => build.txt.4 OK!
Find the output
ll /home/xs32/work/CLEO/analysis/DHad/tmp -rw-r--r-- 1 xs32 cms 30K Feb 11 10:21 data31_dskim_evtstore.root
Open with ROOT, OK.
Reply to HN … sent.
Ready to process 281/pb.
Generate Signal MC (2.0)
First Try
-
Check the webpage .
https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/CLEOcMC
https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/CLEOcSignalMC
- Generate the signal MC D0 > K pi+ against generic D0-bar.
- Create runmc.scr
- Create cleogddbar.tcl
- Create mcpass2ddbar.tcl
-
Create User decay file (ddbar.dec) used in the cleog script above
Put the decay file in the tcsh dir for now.
-
Run it.
./run_mc.src ...
Connection to lnx7228.lns.cornell.edu closed. (exit in the last line of the script)
Need to ask which machine to use for mcgen. … sent.
Use lnx224 for now.
No events processed => run1.log
Ask on CLEOG HN… OK. (not found.)
Ask on BugFix HN and cc to DHad group … sent.
Got help from Laura, need to specify the directory of the decay file.
Change it and run again …
This time, there is the output
run_200978_mcp2.pds, but still error in the output.Change the
run_$MCRUN_cg.logtorun_$env(MCRUN)_cg.logChange the
run_$MCRUN_mcp2.logtorun_$env(MCRUN)_mcp2.logRun again. … Not work. change to back.
Register the log file => run.log
Report on HN … sent.
Message from Laura:
So my MC generation code is a bit complicated. I submit files using:
/home/ljf26/analysis/signalMC/submit_mc.scr The script that actually calls cleog and mcpass2 is /home/ljf26/workshop/DtoPiLNuProc/Class/gen_anyMC.scr These are probably a lot more complicated than you need, but they will give you a general idea of what you need to do, and they did work a few months ago at least. Also, my analysis processor is in /home/ljf26/workshop/DtoPilnuProc/class/DtoPilnuProc.cc. It is very expansive and messy, but the variable isMC is the boolean that extract to find out if I'm using data or MC.
Message from Pete:
/cdat/rd5/cleo3/Offline/rel/20071023_MCP2/src/include/StorageManagement/SMStorageHelper.h:70: T* SMStorageHelper<T>::deliver(SMSourceStream&, unsigned int) [with T = MCParticle]: Assertion `(iVersion-kFirstVersionNumber) < m_deliverers.size()' failed. Are you using the MCGEN release 20071207_MCGEN or newer MCGEN? If you are (and I think you are), you need to use a MCP2 release with the suffix "_A_1". See the end of the "Library release used" section on the followinf wikipage: https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/CLEOcMCstatus
Check with
c3rel, saw20071023_MCP2_A.Try this one…. => run1.log
Sent reply.
Message from Surik:
Try to use absolute path when you are redirecting the suez output. At this point your are using just file name.
Message from Sean:
To get those log files, you might try specifying the absolute path of the log file you're writing to, e.g. in your run_mc.scr, doing suez -f /home/xs32/work/CLEO/analysis/DHad/script/tcl/cleog_ddbar.tcl >& /home/xs32/work/CLEO/analysis/DHad/script/tcl/run_$MCRUN_cg.log (or whatever directory you want to output into)
Try the absolute path … same problem.
Try to echo the MCRUN variable before suez…
echo "test" > run_$MCRUN_cg.log => not work. echo "test" > $MCRUN_cg.log => not work. echo "test" > $MCRUN.log => OK.
Reply to HN… sent.
Message from Sean:
Yes, try using /cdat/tem/xs32/cleo/run_${MCRUN}_cg.log instead - I think that will interpret the variable correctly. (Though any wandering shell-scripting gurus can correct me if I'm wrong).Try this one …
echo "test" > run_${MCRUN}_cg.log OK.Try the run file … done.
Save the log file and pds file … OK.
run200978cg.log, run200978mcp2.log
Send back, mention the change of the Twiki page and raise the question of using two different output dir for the pds file. … sent.
Message from Surik:
Since you find the real cause, I am wondering if you do not specify absolute path for your log files in suez command lines, do you find your log files in the current directory. You have "cd" command line before the suez command line. I am just curious,
Try the relative path … OK.
Reply to Surik. …
-
Ask Peter for the exact Release number for previous Signal MC generation
Look at the dir
/nfs/cor/user/ponyisi/daf9/first … not able to tell.Send email … sent.
Message from Peter:
You can find my MC generation scripts in ~ponyisi/cleog/ ; in particular ~ponyisi/cleog/scripts-summerconf-photosint contains scripts for generating MC with the PHOTOS interference on. The top-level script is fire_up_jobs_new.sh; it should be fairly clear what it does if you read it (makes more sense than most of my code!) The releases are defined in that file; they appear to have been 20050525_MCGEN and 20041104_MCP2. This *should* be mentioned in the CBX, but I might have forgot to put this in.
Need to reply … sent.
Reproduce DtoKpi MC
CLEOG
From the top file fire_up_jobs_new.sh
mkdir $dhad/test mkdir $dhad/test/script cp ~ponyisi/cleog/scripts-summerconf-photosint/fire_up_jobs_new.sh $dhad/test/script cp ~ponyisi/cleog/scripts-summerconf-photosint/test_mode_list $dhad/test/script cp ~ponyisi/cleog/scripts-summerconf-photosint/simple-cleog-generic-array.sh $dhad/test/script
mkdir $dhad/test/script/tag_numbers cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_numbers/number_of_jobs $dhad/test/script/tag_numbers cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_numbers/Single_D0_to_Kpi $dhad/test/script/tag_numbers
mkdir $dhad/test/script/tag_decfiles cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_decfiles/Single_D0_to_Kpi.dec $dhad/test/script/tag_decfiles cp ~ponyisi/cleog/genmc_anders.tcl $dhad/test/script
Edit the fire_up_jobs_new.sh as fire_up_job_DtoKpi.sh
Try the cleog first.
. ./fire_up_job_DtoKpi.sh
Your job-array 1185697.1-10:1 ("cleog-Single_D0_to_Kpi Single_D0_to_Kpi") has been submitted
Check the qsub status … done.
Change the Email address.
. ./fire_up_job_DtoKpi.sh
Your job-array 1185698.1-10:1 ("cleog-Single_D0_to_Kpi Single_D0_to_Kpi") has been submitted
From the output:
nl: /home/xs32/work/CLEO/analysis/DHad/test/script/runlist: No such file or directory ls: /cdat/lns150/disk2/c3mc/RandomTriggerEvents/data*/: Input/output error
Fix the runlist:
cp ~ponyisi/cleog/scripts-summerconf-photosint/runlist $dhad/test/script/
Try again:
. ./fire_up_job_DtoKpi.sh
Your job-array 1185699.1-10:1 ("cleog-Single_D0_to_Kpi Single_D0_to_Kpi") has been submitted
Same output.
Remove the output and try again.
EvtGen:Could not open /home/xs32/work/CLEO/analysis/DHad/test/script/tag_decfiles/Single_D0_to_Kpi.dec
Copy the dec file again:
mkdir $dhad/test/script/tag_decfiles cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_decfiles/Single_D0_to_Kpi.dec $dhad/test/script/tag_decfiles
Try again:
. ./fire_up_job_DtoKpi.sh
Your job-array 1185702.1-10:1 ("cleog-Single_D0_to_Kpi Single_D0_to_Kpi") has been submitted
Check the output message … noticed:
========================== PHOTOS, Version: 2. 0 Released at: 16/11/93 ==========================
%% ERROR-DataStorage.dataStringTagsToDataKeysUsingRecordContents: unknown TypeTag( FATable<CalibratedSVRphiHit> )
%% WARNING-MCInfo.MCDecayMode: Potential Energy nonconservation: vpho --> psi(3770) gamma %% WARNING-MCInfo.MCParticleProperty: Adding a decay mode with a branching fraction of zero!!!! mode 41: frac = 0 D+ --> anti-K0 pi+ pi+ pi- (Model 0)
%% WARNING-MCInfo.MCDecayMode: Potential Energy nonconservation: B_c+ --> anti-B_s0 tau+ anti-nu_tau %% WARNING-MCInfo.MCDecayMode: Potential Energy nonconservation: B_c- --> B_s0 tau- nu_tau %% WARNING-MCInfo.MCParticleProperty: Adding a decay mode with a branching fraction of zero!!!! mode 12: frac = 0 psi(4040) --> D0 anti-D0 pi0 (Model 0)
%% WARNING-wtk_drift_stereo: disc <0. Axial geometry is used. Drift dist, cm = 0.567896426 %% WARNING-check_geant_hit: Illegal Ion Distance = 0.0212069992 %% WARNING-MCResponse.MCCathodesResponseProxy: Too many anode responses %% WARNING-CDOffCal.DataDriftFunction: inverse velocity has illegal value %% WARNING-check_geant_hit: Illegal sinXangle = 1.02591395 restricted
***** Summary Info ***** Stream physics : 1 Stream beginrun : 1 Stream endrun : 1 Stream event : 3137 Stream startrun : 1 Processed 3141 stops.
Find out the output pds file … OK.
Need to ask the line … OK.
ll /home/xs32/work/CLEO/analysis/DHad/test/cleog_0214 total 1.1G -rw-r--r-- 1 xs32 cms 113M Feb 10 17:14 cleog_Single_D0_to_Kpi_10.pds -rw-r--r-- 1 xs32 cms 104M Feb 10 17:21 cleog_Single_D0_to_Kpi_1.pds -rw-r--r-- 1 xs32 cms 104M Feb 10 17:23 cleog_Single_D0_to_Kpi_2.pds -rw-r--r-- 1 xs32 cms 104M Feb 10 17:24 cleog_Single_D0_to_Kpi_3.pds -rw-r--r-- 1 xs32 cms 109M Feb 10 17:21 cleog_Single_D0_to_Kpi_4.pds -rw-r--r-- 1 xs32 cms 110M Feb 10 17:21 cleog_Single_D0_to_Kpi_5.pds -rw-r--r-- 1 xs32 cms 111M Feb 10 17:21 cleog_Single_D0_to_Kpi_6.pds -rw-r--r-- 1 xs32 cms 114M Feb 10 17:13 cleog_Single_D0_to_Kpi_7.pds -rw-r--r-- 1 xs32 cms 115M Feb 10 17:13 cleog_Single_D0_to_Kpi_8.pds -rw-r--r-- 1 xs32 cms 119M Feb 10 17:14 cleog_Single_D0_to_Kpi_9.pds
Ask Peter about the noticed error and the below reason … sent.
ls -R /cdat/lns150/disk2/c3mc/RandomTriggerEvents/data*/ > /dev/null
Message from Peter:
It looks fine. The strange line you noticed was to ensure that the random events files were available (if there's an NFS glitch they may not appear and the job will fail). However they have probably moved them to a different directory and the path I use is wrong. You can remove that line.
PASS2
cp ~ponyisi/cleog/scripts-summerconf-photosint/simple-pass2-generic-array.sh $dhad/test/script cp ~ponyisi/cleog/mcp2_anders.tcl $dhad/test/script cd /home/xs32/work/CLEO/analysis/DHad/test/script/ . ./fire_up_job_DtoKpi.sh
Change the Email !!!
Change the mcp2_anders.tcl:
set histout /home/xs32/work/CLEO/analysis/DHad/test/hist$env(BATCH).rzn
Change the simple-pass2-generic-array.sh, comment out #cd ~/cleog
Add a log output for pass2: => Not work.
#$ -o /home/xs32/work/CLEO/analysis/DHad/test/pass2.txt
Use log as command line.
suez -f mcp2_anders.tcl >& /home/xs32/work/CLEO/analysis/DHad/test/log/pass2_${SGE_TASK_ID}.log
Test … => pass21.log :
%% SYSTEM-Interpreter.TclInterpreter: Error opening file mcp2_anders.tcl
Need to use cd $SCRIPTDIR !
Test … => pass21.log.1 Noticed parts:
%% NOTICE-ConstantsPhase2Delivery.DBCP2Proxy: DBMUHomogenousValues using version 1.1 %% NOTICE-ConstantsPhase2Delivery.DBCP2Proxy: DBMUPoissonValues using version 1.1 *** S/R ERPROP IERR = 2 *** Error in subr. TRPROP 2 called bysubr. ERPROP *** S/R ERPROP IERR = 2 *** Error in subr. TRPROP 2 called bysubr. ERPROP
NTUPLE
– => 0:16
cp ~ponyisi/cleog/scripts-summerconf-photosint/simple-dtuple-generic-array.sh $dhad/test/script
Change:
-
Edit the content of the
local_setupinto the same sh file. - Add log output from the suez .
- Set the event to be 10 in the tcl.
Test … => signalD0toKpi.log
%% SYSTEM-Interpreter.TclInterpreter: Error opening file HadronicDNtupleProc/Test/loadHadronicDNtupleProc.tcl
Use the absolute path… => signalD0toKpi.log.1
%% SYSTEM-Interpreter.TclInterpreter: Error opening file /home/xs32/work/CLEO/analysis/DHad/test/script/tmp/dt-Single_D0_to_Kpi
Make the tmp dir and try again … => signalD0toKpi.log.2
%% ERROR-DynamicLoader.DLSharedObjectHandler: /home/xs32/tmp/build/20050316_FULL/Linux/shlib/MCBeamEnergyFromMCBeamParametersProd.so_20050316_FULL: cannot open shared object file: No such file or directory %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load /home/xs32/tmp/build/20050316_FULL/Linux/shlib/MCBeamEnergyFromMCBeamParametersProd.
Need the lib: MCBeamEnergyFromMCBeamParametersProd
Check the CBX … Not found.
Ask Peter which version to check out and which release to compile. … sent.
Message from Peter:
It's probably fine for you to copy it from /nfs/cor/user/ponyisi/daf9/2005-3/ and compile it in 20050316_FULL.
Copy :
cp -r /nfs/cor/user/ponyisi/daf9/2005-3/MCBeamEnergyFromMCBeamParametersProd $dhad/src/
On lnx134:
Change the setup.sh : c3rel 20050316_FULL
setdhad cd $dhad/src/ c3make
Compile OK.
Try again … => signalD0toKpi.log.3 OK.
Do the full sample (go) … done.
Produce D0BtoKpi
-
CLEOG
cp fire_up_job_DtoKpi.sh gen_Single_D0B_to_Kpi.sh
Change
FILELISTINGS="Single_D0B_to_Kpi"
Provide
Single_D0B_to_Kpifile.Start job … => cleog-SingleD0BtoKpi-1.txt Not work.
Error:
cat: /home/xs32/work/CLEO/analysis/DHad/test/script/tag_numbers/Single_D0B_to_Kpi: No such file or directory
Copy the needed file:
cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_numbers/Single_D0B_to_Kpi $dhad/test/script/tag_numbers cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_decfiles/Single_D0B_to_Kpi.dec $dhad/test/script/tag_decfiles
Try again … => cleog-SingleD0BtoKpi-1.txt OK.
-
PASS2
Edit
gen_Single_D0B_to_Kpi.sh: DOPASS2=1Run … done.
-
NTUPLE
Edit
gen_Single_D0B_to_Kpi.sh: DONTUPLE=1Run … done.
Gen all Siganl MC
-
Ask Brian where to put the big files … sent.
Got dir at :
/nfs/cor/an2/xs32Make softlink
ln -s /nfs/cor/an2/xs32 ~/disk/2 mkdir ~/disk/2/cleo mkdir ~/disk/2/cleo/dhad mv dat ~/disk/2/cleo/dhad/ ln -s ~/disk/2/cleo/dhad/dat $dhad
-
Start the script for CLEOG
Goal:
dhad gen cleog
Start the script in 2.0
cd /home/xs32/work/CLEO/analysis/DHad/src/2.0/python
With initial tag: V02-00-00
cvs tag V02-00-00
Re-direct the dhad command. Linked to dhad-2.0.
Do one mode first:
dhad gen cleog -m 0
Pause for now. Get the generation running first!
-
Generate all of the signal modes
cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_numbers/* $dhad/src/2.0/gen/tag_numbers cp ~ponyisi/cleog/scripts-summerconf-photosint/tag_decfiles/* $dhad/src/2.0/gen/tag_decfiles
Cleog … done.
cd $dhad/src/2.0/gen . ./generate_mc.sh
Qstat with one exception:
1187664 0.55009 cleog-Sing xs32 Eqw 02/12/2009 17:26:31 1 4
Ask on HN for how to re-submit …
Message from Surik:
To learn why it is in Eqw state, you could do "qstat -j JobId" (I in your case JobId is 1187664). If you find that that there is no error from user side, then you could clear error state and resubmit it by saying: qmod -cj JobId.
qstat -j 1187664 => jobstat.txt
Message from Chul Su:
qmod -c 1187664 man qmod
Job-array task 1187664.4 is not in error state
Message from Laura (forward from dlk):
Because of the &%#@* "network glitch", it is fairly common to have a batch job put into Eqw status through no fault of your own: 1060079 0.56000 drawdists_ ljf26 Eqw 10/05/2008 22:37:58 This note is about how to clear that status without deleting and resubmitting that job. Do a qstat -j 1060079 (Queue status for the job 1060079) and look through the voluminous output for the line describing the cause of the error: error reason 1: 10/05/2008 22:38:05 [1648:17407]: error: can't chdir to /home/ljf26: No such file or directory This obviously absurd contention is because there was a temporary dropout of the network connection between the queuing master server and the /home disk. (Sometimes if you write the output to the /tem disk, batch can't find that.) You can clear the error and get a retry by doing qmod -c 1060079 (Queue modify job 1060079 by clearing error status)
Pass2 … done.
Ntuple … done.
Fix the three channels
Edit fixed_single_mode_list. with three channels.
Edit generate_mc.sh for cleog.
CLEOg …
cat: /home/xs32/work/CLEO/analysis/DHad/src/2.0/gen/tag_numbers/Dp_to_Kspipipi: No such file or directory
Add "Single" in the mode list. Re-run. … done.
PASS2 … done.
Still one left:
xs32_lnx224% ll /home/xs32/work/CLEO/analysis/DHad/dat/signal/2.0/cleog_0214/ total 191M -rw-r--r-- 1 xs32 cms 172M Feb 12 20:09 cleog_Single_Dm_to_Kspipipi_1.pds -rw-r--r-- 1 xs32 cms 20M Feb 18 10:59 cleog_Single_Dp_to_Kspipi0_4.pds
xs32_lnx224% tail /home/xs32/work/CLEO/analysis/DHad/dat/signal/2.0/log_c_0525_p2_1104/cleog-Single_Dp_to_Kspipi0-4.txt >> Wed Feb 18 10:59:39 2009 Run: 206080 Event: 460 Stop: event << %% NOTICE-Processor.MCRunEvtNumberProc: RandomGenerator seeds=546597654, 176706065 %% NOTICE-Processor.RunEventNumberProc: Run: 206080 Event: 460 %% NOTICE-Processor.RunEventNumberProc: Run: 206080 Event: 25718 >> Wed Feb 18 10:59:40 2009 Run: 206080 Event: 461 Stop: event << %% NOTICE-Processor.MCRunEvtNumberProc: RandomGenerator seeds=1605634292, 349285818 %% NOTICE-Processor.RunEventNumberProc: Run: 206080 Event: 461 /nfs/cleo3/Offline/rel/20050525_MCGEN/bin/Linux/g++/suez: line 317: 4465 Floating point exception(core dumped) $ECHO $exe $SCRIPT $options Wed Feb 18 10:59:42 EST 2009
Notice the cleog_Single_Dm_to_Kspipipi_1.pds, which cause the 4%
difference. The cleog looked OK. Need to re-run the pass2.
Need to address these two later … Proceed with Ntuple first.
Ntuple … done.
Get yield:
Kpipi0: mode 1 Kspipi0: mode 203 Kspipipi: mode 204 dhad yield regular2 -t s -m 1 dhad yield regular2 -t s -m 203 dhad yield regular2 -t s -m 204
dhad fit regular2 -t s -m 1 --qsub
dhad fit regular2 -t s -m 203 --qsub $
dhad fit regular2 -t s -m 204 --qsub $
Move on to new sample and fix them.
Generate Signal MC with PHOTOS-2.15 (2.1)
First Try
Use 20050417_FULL
dhad yield test -t s --sign 1
Need to specify the root file.
cp $dhad/test/dtuple_c_0525_p2_1104/Single_D0_to_Kpi.root $dhad/dat/signal/2.0/test
Now, just backup the old link and use the new as test.
dhad yield test -t s -m 1 --sign 1
Error:
Error in <TFile::TFile>: file /home/xs32/work/CLEO/analysis/DHad/7.06/dat/signal/test/Single_D0_to_Kpipi0.root does not exist
Need to produce more signal MC … Used the wrong mode number!
dhad yield dtuple_c_0525_p2_1104 -t s -m 0 OK.
Fit for yield:
dhad fit dtuple_c_0525_p2_1104 -t s -m 0
Compare the yield with previous signal MC:
dhad table compare yields dtuple_c_0525_p2_1104 -t s -m 0
Consider using the previous framework to reduce non-essential work …
cd /home/xs32/work/CLEO/analysis/DHad/7.06/dat/signal ln -s /home/xs32/work/CLEO/analysis/DHad/dat/signal/2.0/dtuple_c_0525_p2_1104/ regular2
Use the $dhad/scripy/python as the main dhad.
dhad yield regular2 -t s -m 0 OK. dhad fit regular2 -t s -m 0 ... processing OK.
Need the qsub.
dhad yield regular2 -t s ... done. dhad fit regular2 -t s --qsub
Set the complete qsub env.
log-2009-02-16 20:24:23 … done.
dhad table compare yields regular2 -t s
=> page.
Need to compare efficiency, or generate same number of events as in CBX.
List the generated numbers:
| Mode | CBX 281/pb | Regular 2 | Regular 3 |
|---|---|---|---|
| D0toKpi | 62740 | 3137x10 | 6274x10 |
| D0BtoKpi | 62740 | 3137x10 | 6274x10 |
| D0toKpipi0 | 197350 | 19735x10 | No change |
| D0BtoKpipi0 | 197350 | 19735x10 | No change |
| D0toKpipipi | 99480 | 9948x10 | No change |
| D0BtoKpipipi | 99480 | 9948x10 | No change |
| DptoKpipi | 79980 | 7998x10 | No change |
| DmtoKpipi | 79980 | 7998x10 | No change |
| DptoKpipipi0 | 73380 | 7338x10 | No change |
| DmtoKpipipi0 | 73380 | 7338x10 | No change |
| DptoKspi | 80000 | 8000x10 | No change |
| DmtoKspi | 80000 | 8000x10 | No change |
| DptoKspipi0 | 57160 | 5716x10 | No change |
| DmtoKspipi0 | 57160 | 5716x10 | No change |
| DptoKspipipi | 39310 | 3931x10 | No change |
| DmtoKspipipi | 39310 | 3931x10 | No change |
| DptoKKpi | 20000 | 2000x10 | No change |
| DmtoKKpi | 20000 | 2000x10 | No change |
Only need to re-generate the first one.
Create fixed_single_mode_list. with only one mode.
CLEOg , PASS2, Ntuple … done.
dhad yield regular2 -t s -m 0 OK. dhad fit regular2 -t s -m 0 --qsub
Make the compare page:
dhad table compare yields regular2 -t s
| Mode | signal diff(%) |
|---|---|
| D0toKpi | 1.08 |
| D0BtoKpi | 1.57 |
| D0toKpipi0 | 0.07 |
| D0BtoKpipi0 | -19.95 |
| D0toKpipipi | 0.72 |
| D0BtoKpipipi | 0.70 |
| DptoKpipi | -0.44 |
| DmtoKpipi | 0.42 |
| DptoKpipipi0 | -0.05 |
| DmtoKpipipi0 | -0.00 |
| DptoKspi | -0.24 |
| DmtoKspi | -0.42 |
| DptoKspipi0 | -12.65 |
| DmtoKspipi0 | -4.19 |
| DptoKspipipi | -10.17 |
| DmtoKspipipi | 0.72 |
| DptoKKpi | -0.94 |
| DmtoKKpi | 0.90 |
Message from Peter:
Are you sure that it's not because some of your jobs crashed? You see different effects in D and Dbar.
Eh, are they all on lnx303? Then there's probably something wrong with the node... sometimes the jobs just don't run and the batch queue kills them for lasting too long. Usually in this case I'd just fully rerun those three channels (i.e. including cleog).
Check :
| cleog-SingleDmtoKspipipi-1.txt | lnx311 |
| cleog-SingleD0BtoKpipi0-5.txt | lnx324 |
| cleog-SingleD0BtoKpipi0-6.txt | lnx65110 |
| pass2-SingleD0BtoKpipi0-5.txt | lnx303 |
| pass2-SingleD0BtoKpipi0-6.txt | lnx303 |
| pass2-SingleDmtoKspipipi-1.txt | lnx323 |
Move on to the new sample.
Produce Latest Photos/Dalitz MC
Find the location of the right code
Email Peter for the production process … sent.
Message from Peter:
> > Could you tell me how to generate the MC with "UPDDALITZ" ? What do you mean? > > Also, where is the place to look for the version of the photos? > > Do you mean 'how can I tell what version of PHOTOS is run in a job'? Look in the logfile from the cleog step; there is a lot of output when PHOTOS is initialized and one of the things it prints is the code version.
From the output of cleog:
******************************************************************************* * * ========================== * PHOTOS, Version: 2. 0 * Released at: 16/11/93 * ========================== * * PHOTOS QED Corrections in Particle Decays * * Monte Carlo Program - by E. Barberio, B. van Eijk and Z. Was * From version 2.0 on - by E.B. and Z.W. * *******************************************************************************
Message from Peter:
The directory is /home/ponyisi/cleog/scripts-summerconf, the script is fire_up_jobs_new_photosnew.sh . This should give you both the new photos and the updated Dalitz plot for Ks pi pi0.
Run on the code
Copy the script:
cp /home/ponyisi/cleog/scripts-summerconf/fire_up_jobs_new_photosnew.sh /home/xs32/work/CLEO/analysis/DHad/src/2.1/gen cp /home/ponyisi/cleog/scripts-summerconf/simple-cleog-generic-array-photosnew.sh /home/xs32/work/CLEO/analysis/DHad/src/2.1/gen
The difference between the photos new and old one is:
cd /nfs/cor/an1/ponyisi/2007-4/ . local_setup
Ask Peter what's the correct way to use that, copy dir or re-build … sent.
Message from Peter:
You're probably ok leaving that directory where it is, but if you run into problems you can do that.
Change the file name (email address).
Copy tag numbers, tag decay files, tcl file, mode list.
cp -r $dhad/src/2.0/gen/tag_numbers $dhad/src/2.1/gen/ cp -r $dhad/src/2.0/gen/tag_decfiles $dhad/src/2.1/gen/ cp $dhad/src/2.0/gen/genmc_anders.tcl $dhad/src/2.1/gen/ cp $dhad/src/2.0/gen/single_mode_list $dhad/src/2.1/gen/
Ran cleog on one channel … Error from CLEOG:
=> cleog-SingleDptoKspipi0-1.txt
no files matched glob pattern "/nfs/cleoc/mc1/RandomTriggerEvents/Links/data1/RandomTriggerEvents_*.pds"
Ask Peter … sent.
Peter pointed out:
nl: /home/xs32/work/CLEO/analysis/DHad/src/2.1/gen/runlist: No such file or directory
Copy that file:
cp $dhad/src/2.0/gen/runlist $dhad/src/2.1/gen/
OK now. PHOTOS, Version: 2.15
Run all CLEOG … OK.
Pass2 …
cp /home/ponyisi/cleog/scripts-summerconf/simple-pass2-generic-array.sh /home/xs32/work/CLEO/analysis/DHad/src/2.1/gen
Save as pass2-generic-array.sh
Change the submit job sh.
Change email address.
cp $dhad/src/2.0/gen/mcp2_anders.tcl $dhad/src/2.1/gen
Found leftover pds from pass2.
xs32_lnx570> pwd /home/xs32/work/CLEO/analysis/DHad/dat/signal/2.1/cleog_0214_photosnew xs32_lnx570> ll total 1.6G -rw-r--r-- 1 xs32 cms 494M Feb 19 20:51 cleog_Single_D0B_to_Kpipipi_6.pds -rw-r--r-- 1 xs32 cms 612M Feb 19 18:04 cleog_Single_D0_to_Kpipi0_8.pds -rw-r--r-- 1 xs32 cms 58M Feb 19 18:06 cleog_Single_Dm_to_Kpipipi0_10.pds -rw-r--r-- 1 xs32 cms 161M Feb 19 20:56 cleog_Single_Dm_to_Kspi_3.pds -rw-r--r-- 1 xs32 cms 298M Feb 19 18:22 cleog_Single_Dp_to_Kspi_8.pds -rw-r--r-- 1 xs32 cms 209K Feb 19 18:56 cleog_Single_Dp_to_Kspipipi_5.pds
Fix the bad pass2 runs
cleog_Single_D0B_to_Kpipipi_6.pds
Check the cleog output log …
cd $dhad/dat/signal/2.1/log_c_0525_photosnew_p2_1104 tail -20 cleog-Single_D0B_to_Kpipipi-6.txt
RandomModule Status: Engine = RanecuEngine, seeds = 356693554, 1339856888 %% INFO-JobControl.SummaryModule: ***** Summary Info ***** Stream physics : 1 Stream beginrun : 1 Stream endrun : 1 Stream event : 9948 Stream startrun : 1 Processed 9952 stops.
tail -20 pass2-Single_D0B_to_Kpipipi-6.txt ***** Summary Info ***** Stream physics : 1 Stream beginrun : 1 Stream endrun : 1 Stream event : 9948 Stream startrun : 1 Processed 9952 stops.
Looks OK.
Need to generate: 9948.
Re-do the pass2 with fix task ID = 6. … done.
The cleog pds file has been removed, fix done.
cleog_Single_D0_to_Kpipi0_8.pds
CLEOG:
tail -20 cleog-Single_D0_to_Kpipi0-8.txt
## new death vertex info: ## interaction type : 31 ## geant Parent Track: 69 ## position : (-132.112471,179.447715,-115.464307) ## geant vtx number : 0 %% ERROR-MCInfo.MCParticle: attempt to add fatal interaction vertex to particle that is already dead. Particle: 60 1 gamma (-0.007845, 0.010756,-0.006829; 0.014962) ---- 42 52 2 43 1 61 1 0 previous decay vertex: 43 1 6 (-132.112471,179.447715,-115.464307) 8.823860e+05: gamma --> e+ new decay vertex: 1 0 31 (-132.112471,179.447715,-115.464307) 8.823860e+05: gamma --> suez.exe: /nfs/cleo3/Offline/rel/20050525_MCGEN/src/MCInfo/Class/MCDecayTree/MCParticle.cc:639: void MCParticle::addVertex(MCVertex*): Assertion `false' failed.
Pass2:
tail -20 pass2-Single_D0_to_Kpipi0-8.txt
***** ERROR in HROUT : Current Directory must be a RZ file : %% INFO-RandomModule.RandomModule: RandomModule Status: Engine = RanecuEngine, seeds = 399089476, 1571312518 %% INFO-JobControl.SummaryModule: ***** Summary Info ***** Stream physics : 1 Stream beginrun : 1 Stream event : 15844 Stream startrun : 1 Processed 15847 stops.
File size 40M smaller.
xs32_lnx570> ls *D0_to_Kpipi0_* -lh -rw-r--r-- 1 xs32 cms 196M Feb 19 23:55 pass2_Single_D0_to_Kpipi0_10.pds -rw-r--r-- 1 xs32 cms 194M Feb 20 00:14 pass2_Single_D0_to_Kpipi0_1.pds -rw-r--r-- 1 xs32 cms 192M Feb 20 00:43 pass2_Single_D0_to_Kpipi0_2.pds -rw-r--r-- 1 xs32 cms 191M Feb 20 00:42 pass2_Single_D0_to_Kpipi0_3.pds -rw-r--r-- 1 xs32 cms 192M Feb 19 23:54 pass2_Single_D0_to_Kpipi0_4.pds -rw-r--r-- 1 xs32 cms 193M Feb 19 23:52 pass2_Single_D0_to_Kpipi0_5.pds -rw-r--r-- 1 xs32 cms 194M Feb 20 00:55 pass2_Single_D0_to_Kpipi0_6.pds -rw-r--r-- 1 xs32 cms 193M Feb 20 00:46 pass2_Single_D0_to_Kpipi0_7.pds -rw-r--r-- 1 xs32 cms 155M Feb 20 00:00 pass2_Single_D0_to_Kpipi0_8.pds -rw-r--r-- 1 xs32 cms 193M Feb 19 23:54 pass2_Single_D0_to_Kpipi0_9.pds
Consult Peter for generating single cleog, efficiency… sent.
Message from Peter:
If the cleog output is still there it means the *pass2* job failed. What is the pass2 log?
Sent reply.
Message from Peter:
Well, in this case it looks like the pass2 file should have been created (though with fewer events than you requested); you should check the actual pass2 pds files.
Sent reply.
Find the supposed producing number: 19735
fix_mc_job.sh … done.
Check the tail:
tail -20 cleog-Single_D0_to_Kpipi0-8.txt
Got the wrong number! Need to be 8!
Re-submit job with Task ID = 8. … done.
tail -20 cleog-Single_D0_to_Kpipi0-8.txt ... OK.
Run pass2 …
Fix the rest
cleog_Single_Dm_to_Kpipipi0_10.pds cleog_Single_Dm_to_Kspi_3.pds cleog_Single_Dp_to_Kspi_8.pds cleog_Single_Dp_to_Kspipipi_5.pds
-
Single_Dm_to_Kpipipi0_10tail -20 cleog-Single_Dm_to_Kpipipi0-10.txt Floating point exception(core dumped) $
Need to redo cleog:
-
Change
fix_mc_job.shDOCLEOG=1, DOPASS2=0, DONTUPLE=0, FIXTASKID=10
-
Change
fix_single_mode_listUse
Single_Dm_to_Kpipipi0 -
Run script
. ./fix_mc_job.sh… done. -
Pass2 …
tail -20 cleog-Single_Dm_to_Kpipipi0-10.txt OK.
dhad gen pass2 Single_Dm_to_Kpipipi0 task 10 ... OK.
-
Change
-
cleog_Single_Dm_to_Kspi_3tail -20 cleog-Single_Dm_to_Kspi-3.txt
STOP BIMPCT statement executed
Re-run cleog:
dhad gen cleog Single_Dm_to_Kspi task 3 ... OK.
pass2:
dhad gen pass2 Single_Dm_to_Kspi task 3 ... done.
-
The rest two
tail -20 cleog-Single_Dp_to_Kspi-8.txt dhad gen cleog Single_Dp_to_Kspi task 8 ... OK dhad gen pass2 Single_Dp_to_Kspi task 8 ... done.
tail -20 cleog-Single_Dp_to_Kspipipi-5.txt File FLUKAAF.DAT not found
dhad gen cleog Single_Dp_to_Kspipipi task 5 ... OK dhad gen pass2 Single_Dp_to_Kspipipi task 5 ... done.
Run Dtuple
cp /home/ponyisi/cleog/scripts-summerconf/simple-dtuple-generic-array.sh $dhad/src/2.1/gen
Save as dtuple-generic-array.sh.
Change email, add mkdir for tmp, run … done.
Compare yields with photos 2.15
Link the root files.
cd $dhad/7.06/dat/signal ln -s /home/xs32/work/CLEO/analysis/DHad/dat/signal/2.1/dtuple_c_0525_photosnew_p2_1104/ regular3
Extract yields.
dhad yield regular3 -t s ... done.
Fit … done.
dhad fit regular3 -t s --qsub
dhad table compare yields regular3 -t s
| Mode | signal diff(%) |
|---|---|
| D0toKpi | 0.31 |
| D0BtoKpi | -0.19 |
| D0toKpipi0 | 0.06 |
| D0BtoKpipi0 | -0.69 |
| D0toKpipipi | 0.56 |
| D0BtoKpipipi | 0.27 |
| DptoKpipi | -0.57 |
| DmtoKpipi | 0.31 |
| DptoKpipipi0 | 0.46 |
| DmtoKpipipi0 | -9.51 |
| DptoKspi | -0.87 |
| DmtoKspi | -0.90 |
| DptoKspipi0 | -0.42 |
| DmtoKspipi0 | 0.44 |
| DptoKspipipi | 1.63 |
| DmtoKspipipi | -0.61 |
| DptoKKpi | -0.34 |
| DmtoKKpi | 1.56 |
Check the source of three modes
DmtoKpipipi0, DptoKspipipi, DmtoKKpi
-
DmtoKpipipi0 => fixed.
CLEOg:
tail -20 $dhad/dat/signal/2.1/log_c_0525_photosnew_p2_1104/cleog-Single_Dm_to_Kpipipi0-1.txt
Job ID Processed Stream event 1 7342 7338 2 7342 7338 3 0 0 Need tool:
dhad check cleog Single_Dm_to_Kpipipi0
JobID Processed Stream event 1 7342 7338 2 7342 7338 3 N/A N/A 4 7342 7338 5 7342 7338 6 7342 7338 7 7342 7338 8 7342 7338 9 7342 7338 10 7342 7338 Rerun cleog:
dhad gen cleog Single_Dm_to_Kpipipi0 task 3 ... done.
Check log:
dhad check cleog Single_Dm_to_Kpipipi0 ...OK now.
Ran pass2:
dhad gen pass2 Single_Dm_to_Kpipipi0 task 3 ... done.
Check log:
dhad check pass2 Single_Dm_to_Kpipipi0 ... OK.
Run dtuple …
dhad gen dtuple Single_Dm_to_Kpipipi0 ... done.
Extact yield:
dhad yield regular3 -t s -m 201
Fit … done.
dhad fit regular3 -t s -m 201 --qsub
Compare again:
dhad table compare yields regular3 -t s
Mode signal diff(%) D0toKpi 0.31 D0BtoKpi -0.19 D0toKpipi0 0.06 D0BtoKpipi0 -0.69 D0toKpipipi 0.56 D0BtoKpipipi 0.27 DptoKpipi -0.57 DmtoKpipi 0.31 DptoKpipipi0 0.42 DmtoKpipipi0 0.48 DptoKspi -0.87 DmtoKspi -0.90 DptoKspipi0 -0.42 DmtoKspipi0 0.44 DptoKspipipi 1.63 DmtoKspipipi -0.61 DptoKKpi -0.34 DmtoKKpi 1.56 Fixed the difference.
http://www.lepp.cornell.edu/~xs32/private/DHad/7.06/tables/compare_yields_regular3.html
Create the DHad Meeting page … https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/DHadGroupMeeting
-
DptoKspipipi
dhad check cleog Single_Dp_to_Kspipipi
JobID Processed Stream event 1 3935 3931 2 3935 3931 3 3935 3931 4 3935 3931 5 3935 3931 6 3935 3931 7 3935 3931 8 3935 3931 9 3935 3931 10 3935 3931 check pass2:
tail -20 $dhad/dat/signal/2.1/log_c_0525_photosnew_p2_1104/pass2-Single_Dp_to_Kspipipi-1.txt
dhad check pass2 Single_Dp_to_Kspipipi
JobID Processed Stream event 1 3935 3931 2 3935 3931 3 3935 3931 4 3935 3931 5 3935 3931 6 3935 3931 7 3935 3931 8 3935 3931 9 3935 3931 10 3935 3931 Looks OK, what might be the reason?
-
DmtoKKpi
dhad check cleog Single_Dm_to_KKpi
JobID Processed Stream event 1 2004 2000 2 2004 2000 3 2004 2000 4 2004 2000 5 2004 2000 6 2004 2000 7 2004 2000 8 2004 2000 9 2004 2000 10 2004 2000 dhad check pass2 Single_Dm_to_KKpi
JobID Processed Stream event 1 2004 2000 2 2004 2000 3 2004 2000 4 2004 2000 5 2004 2000 6 2004 2000 7 2004 2000 8 2004 2000 9 2004 2000 10 2004 2000 Look OK. Need to discuss.
Add error to the difference
Refer to table double_generic_eff
dhad-2.1 table compare yields signal regular3
Where σ = (a-b)/σb
Also add the original columns OK. => Table link.
Add more digits to the table
dhad-2.1 table compare yields signal regular3 --set rnd=0.01
fix in the tabletools.py:
c_err = a_val/float(b_err)
Now, one digit is OK.
Update the table with two digits. => Signal Table
Update the data compare table:=> Data Table
dhad-2.1 table compare yields data regular3 --set rnd=0.001
Generate double tag signal MC (2.1) [%]
Setup env
cd $dhad/src/2.1/gen cp ~ponyisi/cleog/scripts-summerconf-photosint/double_mode_list .
dhad-2.1 gen cleog Single_Dm_to_Kspi task 1 --test OK.
dhad-2.1 gen cleog Double_Dp_to_Kspipipi__Dm_to_KKpi task 1 --test OK.
Test 10 events. (cleog-generic-array.sh)
dhad-2.1 gen cleog Double_Dp_to_Kspipipi__Dm_to_KKpi task 1 OK.
Use the standard number of events… OK.
dhad-2.1 gen cleog Double_Dp_to_Kspipipi__Dm_to_KKpi task 1-10 --test OK.
Generate the events
dhad-2.1 gen cleog Double_Dp_to_Kspipipi__Dm_to_KKpi task 1-10 dhad-2.1 check cleog Double_Dp_to_Kspipipi__Dm_to_KKpi OK.
dhad-2.1 gen pass2 Double_Dp_to_Kspipipi__Dm_to_KKpi task 1-5 --test OK. dhad-2.1 gen pass2 Double_Dp_to_Kspipipi__Dm_to_KKpi task 1-10 dhad-2.1 check pass2 Double_Dp_to_Kspipipi__Dm_to_KKpi OK.
dhad-2.1 gen dtuple Double_Dp_to_Kspipipi__Dm_to_KKpi dhad-2.1 check dtuple Double_Dp_to_Kspipipi__Dm_to_KKpi OK.
Process 281 data (2.1)
list the needed data set
ls /home/xs32/work/CLEO/analysis/DHad/7.06/dat/data/ data31_dskim_evtstore.root data32_dskim_evtstore.root data33_dskim_evtstore.root data35_dskim_evtstore.root data36_dskim_evtstore.root data37_dskim_evtstore_1.root data37_dskim_evtstore.root
data 31
cp $dhad/test/script/test_data31.sh $dhad/src/2.1/gen
Edit it according to dtuple-generic-array.sh.
Test run for 10 evts … OK.
Run dtuple-data31.sh. … done.
Comment out the WIDEKS=1, save them in the dir wideks.
Then re-run , submitted …
data 32
cp /nfs/cor/user/ponyisi/daf9/simple-dtuple-data32.sh $dhad/src/2.1/gen
Edit according to dtuple-data31.sh and save as dtuple-data32.sh .
Submit … OK.
data 33
cp /nfs/cor/user/ponyisi/daf9/simple-dtuple-data33.sh $dhad/src/2.1/gen
Copy 32 to 33, ask Peter about the local setup duplication … OK to comment out.
Submit …
data 35
cp /nfs/cor/user/ponyisi/daf9/simple-dtuple-data35.sh $dhad/src/2.1/gen
Copy 33 to 35.
submit …
data 36
Copy 35 to 36, edit.
cp /nfs/cor/user/ponyisi/daf9/simple-dtuple-data36.sh $dhad/src/2.1/gen
submit…
data 37
Copy 36 to 37, edit.
cp /nfs/cor/user/ponyisi/daf9/simple-dtuple-data37.sh $dhad/src/2.1/gen
submit …
Extract yields
Link the root files to 7.06/dat/data/regular3
dhad yield regular3 -t d -m 0 OK.
dhad yield regular3 -t d --qsub ... done.
Fit data
Set up the 2.1 env:
cd /home/xs32/work/CLEO/analysis/DHad/src/2.1 cvs co -r V02-01-00 -d python dhad/src/python . /home/xs32/work/CLEO/analysis/DHad/src/2.1/bash/dhadenv.sh
dhad fit regular3 -t d -m 0 Not working!
Use the script/python to cover the head of src/python :
cvs co -d python dhad/src/python cp /home/xs32/work/CLEO/analysis/DHad/script/python/* /home/xs32/work/CLEO/analysis/DHad/src/2.1/python cvs up dhad fit regular3 -t d -m 0 OK.
Check in the code with tag V02-01-01 OK.
Edit the qsub with one line:
. /home/xs32/work/CLEO/analysis/DHad/src/2.1/bash/dhadenv.sh
Try one mode:
dhad fit regular3 -t d -m 0 --qsub
Not work.
Add one line : export ROOTLIB=$ROOTSYS/lib
dhad fit regular3 -t d -m 0 --qsub
log-2009-03-03 14:24:41 Same problem.
Use the full setup
dhad fit regular3 -t d -m 0 --qsub
log-2009-03-03 15:35:00 same problem.
Try signal fit:
dhad fit regular3 -t s -m 0 --qsub
log-2009-03-03 15:41:39 same problem. This should be caused by the script.
Use the script/python code:
dhad-2.1 fit regular3 -t d -m 0 --qsub
For other modes:
setdhad dhadrel 2.1 dhad-2.1 fit regular3 -t d -m 1 OK.
dhad-2.1 fit regular3 -t d -m 1 --qsub
log-2009-03-04 11:27:13 working.
dhad-2.1 fit regular3 -t d -m 3 --qsub
dhad-2.1 fit regular3 -t d -m 200 --qsub
dhad-2.1 fit regular3 -t d -m 201 --qsub
dhad-2.1 fit regular3 -t d -m 202 --qsub
dhad-2.1 fit regular3 -t d -m 203 --qsub
dhad-2.1 fit regular3 -t d -m 204 --qsub
dhad-2.1 fit regular3 -t d -m 205 --qsub
Compare yields
dhadrel 2.1 dhad-2 table compare yields regular3 -t d -m 0
Need to finish the other modes. OK.
dhad-2.1 table compare yields data regular3
Process 818 data (2.2) [88%]
First Try on data 43,44,45,46
https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/DatasetSummary
data 43,44,45,46
-
Data 43
Create dtuple-data43.sh
qsub dtuple-data43.sh ...
-
Data 44
qsub dtuple-data44.sh ...
-
Data 45
qsub dtuple-data45.sh ...
-
Data 46
qsub dtuple-data46.sh ...
Error :
%% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: can't read "preliminaryPass2": no such variable
Save the output => data43.txt
Ask Peter … sent.
Message from Peter:
You must edit $env(USER_SRC)/HadronicDNtupleProc/Test/dataselection.tcl to include a section for 'data43_dskim_evtstore'.
Set up src 2.2
-
Branch out SRC python B02-01
cd /home/xs32/work/CLEO/analysis/DHad/src/2.1/python cvs tag V02-01-02 cvs tag -b -r V02-01-02 B02-01 cvs up -r B02-01 OK.
-
Create src/2.2 based on V02-01-02
cd $dhad/src cvs co -d 2.2 -r V02-01-02 dhad/src cp ../2.1/* -r . cd python cvs tag -r V02-01-02 -b B02-02 cvs up -r B02-02
-
Check out the cleo code
cd $dhad/src/2.2/cleo export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co -rv02_00_04 MCCCTagger cleo3cvs co -rponyisi060929 HadronicDNtupleProc cleo3cvs co CWNFramework
Log on lnx134
setdhad 2.2 cd $dhad/src/2.2/cleo c3make ... OK.
-
Test run on data31
Edit dtuple-data31.sh
mkdir /home/xs32/work/CLEO/analysis/DHad/dat/data/$rel cd $dhad/src/2.2/gen . dtuple-data31.sh
Error:
%% ERROR-DynamicLoader.DLSharedObjectHandler: /nfs/cleo3/Offline/rel/20050417_FULL/other_sources/lib/Linux/g++ /libXrdPosix.so: undefined symbol: XrdSecGetProtocol
Compile again … Use the previous env setup… OK
Second Try : Encounter the coredump
-
Edit dataselection.tcl
Compare with the new one
cd src/2.2/cleo mkdir new cd new export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co HadronicDNtupleProc
No data43 info in the tcl.
Add section in the tcl:
if { ( $env(INPUTDATA) == "data43_dskim_evtstore" ) } { set skim yes set preliminaryPass2 no set millionMC no set mc no set use_setup_analysis yes module sel EventStoreModule eventstore in 20050429 dtag all dataset data43 }Ask Peter for comfirm with the date … sent.
Message from Peter:
Don't know if you should keep the same timestamp - in fact, probably not, I don't think the data was even collected then?
-
Try it for 1 event
Edit the data43 sh file
Edit the tcl file for one event. Error:
%% SYSTEM-Module: added command "eventstore" from EventStoreModule %% SYSTEM-MySQLQuerier.MySQLQuerier: Connecting to EventStore@lnx150.lns.cornell.edu %% ERROR-EventStoreModuleBase.EventStoreModuleBase: no runs found in required range
-
Use Dskim 20070822
From page:
https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/FederationDetails
Change the tcl.
eventstore in 20070822 dtag all dataset data43
OK. Got output.
Reset the tcl to run for all events.
qsub dtuple-data43.sh
Notice error in the data31 (dtuple-data31.txt)
Suez.setup_analysis_command...> error "setup_analysis: unknown option: $subCommand" %% ERROR-Processor.RunEventNumberProc: No data of type "DBRunHeader" "" "" in Record beginrun Please add a Source or Producer to your job which can deliver this data.
Try to checkout:
cd src/2.2/cleo export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co DBRunHeaderProd c3make
Still has the same problem.
Ask Peter … sent.
Re-check the previous data 281/pb process, it had the same error message in the log file.
Sent to Peter.
Finished processing the data43 => dtuple-data43.txt
Notice lines:
>> Wed Jun 3 15:48:45 2009 Run: 221242 Event: 4599 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of DD candidates exceeded >> Wed Jun 3 16:12:07 2009 Run: 221242 Event: 6470 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: To many missing masses %% NOTICE-Processor.HadronicDNtupleProc: To many missing masses
No summary info at the bottom either.
Post on DHad HN … sent.
Message from Anders:
Have you looked at why this message gets produced? It means that you try to produce to many missing mass candidates which are used for tracking efficiency studies etc. I would have hoped that the code would have handled this gracefully, but there seems to be some problem as you don't get any further output. Do you get any error output from the job?
Respond with the core dump.
Message from Anders:
are they from the jobs you submitted? (e.g. by looking at the timestamps?) If so can you get a trace back and see where it failed.
Check the date stamp.
ls -l /home/xs32/work/CLEO/analysis/DHad/dat/data/2.2/ Jun 3 15:47 data43_dskim_evtstore.root Jun 4 15:26 dtuple-data43.txt => this time is coused by the latest data44-46 run. Resubmited.
ls -l /home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/ Jun 3 17:02 core.2694 Jun 3 17:02 core.2718 Jun 3 17:02 core.2738
Open another section for tracing the core dump …
-
Run on data44-46
Add sections in the tcl file.
Edit dtuple-data44.sh, 46, 46. Remember to change the log file!!
qsub dtuple-data44.sh qsub dtuple-data45.sh qsub dtuple-data46.sh
Data44 finished, but the log file has the same error at the end => dtuple-data44.txt
Trace the coredump.
Ask Peter about running the 818 data… later…
Trace the coredump: CPU time limit exceeded
Start from page: http://cs.baylor.edu/~donahoo/tools/gdb/tutorial.html
cd /home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/ gdb core.2694 "/a/lnx112/nfs/cor/user/xs32/local/work/CLEO/analysis/DHad /src/2.2/cleo/core.2694": not in executable format: File format not recognized
Respond to Anders … sent.
Message from Anders:
to use the debugger you need to do: gdb <path to the suez executable> core.2694
From the cleo3defs:
suez() { ${C3_LIB}/bin/${OS_NAME}/${C3CXXTYPE}/suez "$@"; }
Redo:
setdhad 2.2
echo ${C3_LIB}/bin/${OS_NAME}/${C3CXXTYPE}
/nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++
gdb /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez core.2694
This GDB was configured as "i386-redhat-linux-gnu"..."/a/lnx134/cdat/rd5/cleo3/Offline/rel/20050417_FULL/bin/Li
nux/g++/suez": not in executable format: File format not recognized
Core was generated by `bash /nfs/sge/root/default/spool/lnx1622/job_scripts/1275637'.
Program terminated with signal 24, CPU time limit exceeded.
#0 0x00b3e7a2 in ?? ()
gdb /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez.exe core.2694
This GDB was configured as "i386-redhat-linux-gnu"...Using host libthread_db library "/lib/tls/libthread_db.so. 1". warning: core file may not match specified executable file. Core was generated by `bash /nfs/sge/root/default/spool/lnx1622/job_scripts/1275637'. Program terminated with signal 24, CPU time limit exceeded. #0 0x00b3e7a2 in ?? ()
gdb /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez.exe core.2718 warning: core file may not match specified executable file. Core was generated by `bash /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez -f HadronicDNtuple'. Program terminated with signal 24, CPU time limit exceeded. #0 0x00b3e7a2 in ?? ()
gdb /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez.exe core.2738 Core was generated by `/nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez.exe -f HadronicDNtupleP'. Program terminated with signal 24, CPU time limit exceeded. Cannot access memory at address 0x48ca6000 #0 0x002739ce in ?? ()
Respond to Anders … sent.
Message from Anders:
Are you actually running out of CPU?
Check the new finished data44 coredump.
gdb /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez.exe core.5477 warning: core file may not match specified executable file. Core was generated by `bash /nfs/sge/root/default/spool/lnx65111/job_scripts/1277459'. Program terminated with signal 24, CPU time limit exceeded. #0 0x003e17a2 in ?? ()
gdb bash /nfs/sge/root/default/spool/lnx65111/job_scripts/1277459 core.5477 /nfs/sge/root/default/spool/lnx65111/job_scripts/1277459: No such file or directory.
gdb /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez core.5499 Core was generated by `bash /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez -f HadronicDNtuple'. gdb bash /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez -f HadronicDNtuple core.5499 "/nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez" is not a core dump: File format not recognized
gdb bash core.5499
Core was generated by `bash /nfs/cleo3/Offline/rel/20050417_FULL/bin/Linux/g++/suez -f HadronicDNtuple'. Program terminated with signal 24, CPU time limit exceeded. warning: .dynamic section for "/lib/libtermcap.so.2" is not at the expected address warning: difference appears to be caused by prelink, adjusting expectations warning: .dynamic section for "/lib/libdl.so.2" is not at the expected address warning: difference appears to be caused by prelink, adjusting expectations warning: .dynamic section for "/lib/tls/libc.so.6" is not at the expected address warning: difference appears to be caused by prelink, adjusting expectations warning: .dynamic section for "/lib/ld-linux.so.2" is not at the expected address warning: difference appears to be caused by prelink, adjusting expectations Reading symbols from /lib/libtermcap.so.2...(no debugging symbols found)...done. Loaded symbols for /lib/libtermcap.so.2 Reading symbols from /lib/libdl.so.2...(no debugging symbols found)...done. Loaded symbols for /lib/libdl.so.2 Reading symbols from /lib/tls/libc.so.6...(no debugging symbols found)...done. Loaded symbols for /lib/tls/libc.so.6 Reading symbols from /lib/ld-linux.so.2... (no debugging symbols found)...done. Loaded symbols for /lib/ld-linux.so.2 #0 0x003e17a2 in _dl_sysinfo_int80 () from /lib/ld-linux.so.2
Respond to Anders … sent.
Message from Anders:
can you send me the code that generated the printouts about to many missing mass candidates.
Found in bottom of HadronicDNtupleProcmissingMass.cc:
if (tuple.nmiss>=10000) {
report( NOTICE, kFacilityString )
<<"To many missing masses"<<endl;
tuple.nmiss=9999;
}
Message from Peter:
Depending on whether or not you want to redo the relevant systematics studies, you may be able to comment out the calls to the missing mass code - this saves a lot of CPU and disk space...
Message from Fan:
Are you running your jobs on the data before DSkim or After DSkim? I guess such kind of problem maybe due to the array exceed. You can check some particular event region to see whether there are some strange events or not. Such as there are many soft photons in one event (over 300 or something like that). This kind of event sometime can cause serious multi-candidate problem and reach your array limit.
Trace the "To many missing masses" problem
Print debug info according to Anders:
$dhad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProcmissingMass.cc
Line 460:
cout << "Number of recoil cands:"<<RecoilList.size()<<endl;
Compile … OK.
cd $dhad/src/2.2/gen . dtuple-data43.sh
Error:
%% ERROR-DynamicLoader.DLSharedObjectHandler: /nfs/cleo3/Offline/rel/20050417_FULL/other_sources/lib/Linux/g++ /libXrdPosix.so: undefined symbol: XrdSecGetProtocol %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load EventStoreModule. %% SYSTEM-Interpreter.TclInterpreter: Tcl_Eval error: ERROR: cannot load EventStoreModule.
Try again.
Same error.
Re-compile … still the same.
Comment out the print out … same error.
Ask on Prelimbugs HN … sent.
Sent service request.
Message from Dan:
https://hypernews.lepp.cornell.edu/HyperNews/get/Prelimbugs/26/1.html:
echo $LD_LIBRARY_PATH print? That error usually means that you have a version of ROOT on your LD_LIBRARY_PATH that is incompatible with the version of the xrootd libraries EventStoreModule now uses. We encourage people to *not* put ROOT on LD_LIBRARY_PATH; use the "c3root" command to run root instead.
Remove the ROOT from the LD lib in setup.sh:
#export LD_LIBRARY_PATH=$C3PYTHONPATH/lib:$ROOTSYS/lib export LD_LIBRARY_PATH=$C3PYTHONPATH/lib
Works.
Submit qsub … => data43.txt
Send to Anders … sent.
Edit from Anders :
Move the print out into line 676:
report( NOTICE, kFacilityString ) << "Number of recoil cands:"<<RecoilList.size()<<endl;
break;
Compile … OK.
Need to break down the data43 into two pieces.
Break down data43 into two
From page: https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/DatasetSummary
| data set | begin | end | runs | first 1000 runs |
| data43 | 220898 | 223271 | 2373 | 221898 |
In this page: https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/EventStore
eventstore in 20090402 runs 202126 203103
Edit the dataselection.tcl:
if { ( $env(INPUTDATA) == "data43_1_dskim_evtstore" ) } {
set skim yes
set preliminaryPass2 no
set millionMC no
set mc no
set use_setup_analysis yes
module sel EventStoreModule
eventstore in 20070822 dtag all runs 220898 221898
}
if { ( $env(INPUTDATA) == "data43_2_dskim_evtstore" ) } {
set skim yes
set preliminaryPass2 no
set millionMC no
set mc no
set use_setup_analysis yes
module sel EventStoreModule
eventstore in 20070822 dtag all runs 221899 223271
}
Create dtuple-data43_1.sh
Test run … OK.
qsub dtuple-data43_1.sh
Log file => dtuple-data43_1.txt
Error happend at:
>> Wed Jun 10 14:14:59 2009 Run: 221242 Event: 4599 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of DD candidates exceeded >> Wed Jun 10 14:32:54 2009 Run: 221242 Event: 6470 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: To many missing masses %% NOTICE-Processor.HadronicDNtupleProc: Number of recoil cands:12611
Find ways to get over this event …
Create dtuple-data43_2.sh
qsub dtuple-data43_2.sh
Log file => dtuple-data43_2.txt
Just stopped at :
>> Wed Jun 10 12:12:52 2009 Run: 221988 Event: 22938 Stop: event << %% NOTICE-Processor.RunEventNumberProc: Run: 221988 Event: 22938
Break the data from here for another sub sample …
Read the log info about the DHad code …
Event Display the trouble event
(Run 221242 Event: 6470)
EventDisplay :
Refer to Suez talk.
Try the example.tcl:
suez -f example.tcl
Can open the display frame. But when click on the "continue", it showed this error message:
Fail to open /cdat/cleo/detector/event/daq/226000/226000/r226000.bin: No such file or directory %% WARNING-Processor.SpExtractShowerAttributesProc.ShowerAttributesList: Fail to open /cdat/cleo/detector/event/daq/226000 /226000/r226000.bin Please immediately send request to service@mail.lns.cornell.edu Either a disk or system serving EventStore data is down
Display the run 221242:
In file HadronicDNtupleProc/Test/dataselection.tcl, eventstore 20070822.
Run the display.tcl, get the same error:
%% INFO-Processor.SpExtractDRHitsProc: setting up CalibratedCathodeHits for display %% WARNING-UInt32StreamsFormat.FDIStream: Attempt to open /cdat/cleo/detector/event/daq/221200/221242/r221242.bin %% WARNING-UInt32StreamsFormat.FDIStream: Attempt to open /cdat/cleo/detector/event/daq/221200/221242/r221242.bin Fail to open /cdat/cleo/detector/event/daq/221200/221242/r221242.bin: No such file or directory %% WARNING-Processor.SpExtractDRHitsProc.CalibratedDRHitList: Fail to open /cdat/cleo/detector/event/daq/221200/221242/r22 1242.bin Please immediately send request to service@mail.lns.cornell.edu Either a disk or system serving EventStore data is down
Ask on Preliminary bugs HN … link.
Reply from Dan:
We only have a fraction of the raw data spinning on disk, usually the data that were most recently processed through pass2. Since we never implemented on-demand staging in eventstore, any raw data has go be pre-staged for use. If there's a short list of runs you want to look at, I can stage them fairly quickly; if there's a long list, you may to negotiate with Anders and Souvik. If you just want to look at something, most runs in data48 are currently staged, along with the first 100 runs of data47.
Ask for this particular Run 221242 Event: 6470 sent.
Reply from Dan:
I've initiated staging of 221242; you can ls /cdat/sol404/disk1/objy/cleo/detector/event/daq/221200/221242/r221242.bin to see when it's ready (I'd expect it within an hour or two unless there are problems with the tape robot).
ls OK.
Suez> goto hlep
goto 221242 6470 go
Save the picture:

Inplement code to bypass the trouble events
Re-run the test to identify trouble events
In HadronicDNtupleProc_missingMass.cc:
if (tuple.nmiss>=10000) {
report( NOTICE, kFacilityString )
<<"To many missing masses"<<endl;
tuple.nmiss=9999;
report( NOTICE, kFacilityString ) << "Number of recoil cands:"<<RecoilList.size()<<endl;
break;
}
Test data43, run 221242
Create $dhad/src/2.2/gendtuple-data43-test.sh :
#$ -o /home/xs32/work/CLEO/analysis/DHad/dat/data/2.2/dtuple-data43-test.txt export INPUTDATA=data43_test_dskim_evtstore export FNAME=data43_test_dskim
Add lines in dataselection.tcl
if { ( $env(INPUTDATA) == "data43_test_dskim_evtstore" ) } {
set skim yes
set preliminaryPass2 no
set millionMC no
set mc no
set use_setup_analysis yes
module sel EventStoreModule
eventstore in 20070822 dtag all runs 221242 221242
Job complete: Log file dtuple-data43-test.txt
Job 1545587 (dtuple-data43-test.sh) Complete User = xs32 Queue = all.q@lnx187.lns.cornell.edu Host = lnx187.lns.cornell.edu Start Time = 10/13/2009 10:44:11 End Time = 10/13/2009 11:12:41 User Time = 00:27:09 System Time = 00:00:08 Wallclock Time = 00:28:30 CPU = 00:27:17 Max vmem = 1.798G Exit Status = 0
Event 794:

In the log file:
>> Tue Oct 13 10:46:43 2009 Run: 221242 Event: 4599 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of DD candidates exceeded
Event 4599:

Fix the Hard limit on number of DD candidates
Change the code:
if( tuple.nddcand >= MAXNDDCAND )
{
report( NOTICE, kFacilityString )
<< "Hard limit on number of DD candidates exceeded" << endl;
continue; // added by xs32
}
Compile …
sl lc setdhad 2.2 cd $dhad/src/2.2/cleo c3make
Error message:
/home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProc.cc: In member function `virtual ActionBase::ActionResult HadronicDNtupleProc::event(Frame&)': /home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProc.cc:921: continue statement not within a loop gmake[1]: *** [HadronicDNtupleProc.o] Error 1
Read basics of event loop in CLEO101.
Read the example code : MyFirstProc.cc
Read Writing a Processor. In slide:

Test on the kFalaiure and then ask …
if( tuple.nddcand >= MAXNDDCAND )
{
report( NOTICE, kFacilityString )
<< "Hard limit on number of DD candidates exceeded" << endl;
return ActionBase::kFailed; // added by xs32
}
Compile … OK.
Clear old log file:
rm /home/xs32/work/CLEO/analysis/DHad/dat/data/2.2/dtuple-data43-test.txt
Test again:
cd $dhad/src/2.2/gen qsub dtuple-data43-test.sh
Save log file: dtuple-data43-test.txt
Job summary:
Job 1546917 (dtuple-data43-test.sh) Complete User = xs32 Queue = all.q@lnx189.lns.cornell.edu Host = lnx189.lns.cornell.edu Start Time = 10/14/2009 08:10:06 End Time = 10/14/2009 08:29:40 User Time = 00:19:18 System Time = 00:00:03 Wallclock Time = 00:19:34 CPU = 00:19:21 Max vmem = 1.794G Exit Status = 0
Time spend on event 4599:
| Test | Job start | Job end | Duration |
|---|---|---|---|
| First | Oct 13 10:46:43 2009 | Oct 13 11:10:34 2009 | 0@ 23' 51" |
| Second | Oct 14 08:12:04 2009 | Oct 14 08:28:43 2009 | 0@ 16' 39" |
Time spend on event 6470:
| Test | Job start | Job end | Duration |
|---|---|---|---|
| First | Oct 13 11:10:34 | Oct 13 11:12:41 | 0@ 2' 7" |
| Second | Oct 14 08:28:43 | Oct 14 08:29:40 | 0@ 0' 57" |
Ask Peter … sent.
Comments from David and Jim after the meeting: Use Bad Event List.
Get the Bad Event List
if ( tuple.run == 221242 && tuple.event == 4599 ) {
report( NOTICE, kFacilityString ) << "Bypassing ... Run: " << tuple.run
<< " Event: " << tuple.event << endl;
return ActionBase::kFailed;
}
OK.
Add event 6470 and continue … OK. => data43-test.txt
| Run | Event |
|---|---|
| 221242 | 4599 |
| 221242 | 6470 |
| 221295 | 19061 |
| 221311 | 22714 |
Test all the test runs in data43 (from 221243 to 223271)
Stuck on 221295 Event: 19061 … fixed.
Run: 221311 Event: 22714 : Spent 16 minutes DD limit … fixed.
Run: 221384 Event: 17807 : Spent 7 minutes DD limit … fixed.
Run: 221413 Event: 47962 : 6 min … not this one.
>> Mon Oct 19 14:04:06 2009 Run: 221413 Event: 47962 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: Bypassing ... Run: 221413 Event: 47962 >> Mon Oct 19 14:09:20 2009 Run: 221413 Event: 54964 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: To many missing masses ... %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of DD candidates exceeded >> Mon Oct 19 14:10:01 2009 Run: 221413 Event: 59012 Stop: event << %% NOTICE-Processor.RunEventNumberProc: Run: 221413 Event: 59012
Run: 221413 Event: 54964 : 6 min … fixed.
Run: 221438 Event: 37151 : 5 min … fixed.
Run: 221477 Event: 57536 : 6 min … fixed.
Run: 221492 Event: 62595 : 4 min … fixed.
Job aborted at Run: 221493 Event: Event: 47146
Continue from 221493 to 221898 …
Job aborted at Run: 221493 Event: 47852 :
Job 1553357 (dtuple-data43-test.sh) Aborted Exit Status = 152 Signal = XCPU User = xs32 Queue = all.q@lnx187.lns.cornell.edu Host = lnx187.lns.cornell.edu Start Time = 10/20/2009 10:54:22 End Time = 10/20/2009 10:56:28 CPU = 00:01:50 Max vmem = 1.866G failed assumedly after job because: job 1553357.1 died through signal XCPU (24)
Run: 221493 Event: 47852 : Aborted … bypassed.
Got error:
ERROR: Suez caught "bad_alloc" exception: "St9bad_alloc"
Bypass the previous run … Run: 221493 Event: 47146 …
Still aborted:
Job 1553374 (dtuple-data43-test.sh) Aborted Exit Status = 152 Signal = XCPU User = xs32 Queue = all.q@lnx187.lns.cornell.edu Host = lnx187.lns.cornell.edu Start Time = 10/20/2009 11:36:06 End Time = 10/20/2009 11:38:11 CPU = 00:01:49 Max vmem = 1.868G failed assumedly after job because: job 1553374.1 died through signal XCPU (24)
Bypass the next event : Run: 221493 Event: 47853 … still aborted.
Shrink the range for the trouble event [47853, 60000] … OK.
[60001, 75516 ]
Run: 221493 Event: 60003, 60017, 60067, 60096, 60102, 60198, 60221, 60282, 60301, 60465, 60479, 60628, 60649, 60653, 60705(not filled) … aborted …
Need to read in a bad run list file to avoid compile every time … Done.
- Start the test2 for the second half of data43 (221899 223271)
Run: 221929 Event: 16197 : 8 min …
Run: 221951 Event: 9909 : 5 min …
Run: 221963 Event: 8017 : 16 min …
Run: 221963 Event: 49934 : 4 min…
Aborted at Run: 221988 Event: 22938 …
Avoid missing mass calc
Comment out the if statement.
if ( tuple.run == 221242 && tuple.event == 4599 ||
tuple.run == 221242 && tuple.event == 6470 ||
tuple.run == 221295 && tuple.event == 19061 ||
tuple.run == 221384 && tuple.event == 17807 ||
tuple.run == 221413 && tuple.event == 54964 ||
tuple.run == 221438 && tuple.event == 37151 ||
tuple.run == 221477 && tuple.event == 57536 ||
tuple.run == 221492 && tuple.event == 62595 ||
tuple.run == 221493 && tuple.event == 47146 ||
tuple.run == 221493 && tuple.event == 47852 ||
tuple.run == 221493 && tuple.event == 60003 ||
tuple.run == 221493 && tuple.event == 60017 ||
tuple.run == 221493 && tuple.event == 60067 ||
tuple.run == 221493 && tuple.event == 60096 ||
tuple.run == 221493 && tuple.event == 60102 ||
tuple.run == 221493 && tuple.event == 60198 ||
tuple.run == 221493 && tuple.event == 60221 ||
tuple.run == 221493 && tuple.event == 60282 ||
tuple.run == 221493 && tuple.event == 60301 ||
tuple.run == 221493 && tuple.event == 60465 ||
tuple.run == 221493 && tuple.event == 60479 ||
tuple.run == 221493 && tuple.event == 60628 ||
tuple.run == 221493 && tuple.event == 60649 ||
tuple.run == 221493 && tuple.event == 60653 ||
// tuple.run == 221493 && ( tuple.event <= 47853 || tuple.event >= 60000) ||
tuple.run == 221493 && ( tuple.event <= 60653 || tuple.event >= 75516) ||
tuple.run > 221493 ||
tuple.run == 221311 && tuple.event == 22714
) {
report( NOTICE, kFacilityString ) << "Bypassing ... Run: " << tuple.run
<< " Event: " << tuple.event << endl;
return ActionBase::kFailed;
}
Comment out the missingMassObjects. … done.
Test on run 221242, change line in dataselection.tcl.
qsub dtuple-data43-test.sh
Save log file data43.txt.
"segmentation violation" error.
Comment out additional lines…
/home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProc.cc:581: ` pionListTmp' undeclared (first use this function)
Move on …
/home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProc.cc: In member function `virtual ActionBase::ActionResult HadronicDNtupleProc::event(Frame&)': /home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProc.cc:585: ` kaonListTmp' undeclared (first use this function) /home/xs32/work/CLEO/analysis/DHad/src/2.2/cleo/HadronicDNtupleProc/Class/HadronicDNtupleProc.cc:585: (Each undeclared identifier is reported only once for each function it appears in.)
Uncomment more … OK.
Test run again … log file data43.txt.1 same error.
Comment the delete lines … not work.
Need to understand more about the code …
Suggestion from Peter:
Comment out the makeMissingMassStage
// makeMissingMassStage1(*iDListItem, *labMomentum); // makeMissingMassStage2(tagDecay, *labMomentum);
Test on run 221242: change line in dataselection.tcl.
qsub dtuple-data43-test.sh
Log file data43.txt.
Still spent 1:30 on Run: 221242 Event: 6470.
Move on from run 221243 to 221494 … OK.
mv $dhad/dat/data/2.2/dtuple-data43-test.txt $dhad/log/2009/1103/data43.txt.1
Check the trouble time, add to EventsToSkip.txt
221295 19061 1hour 221493 61358 1:30
Test 221295 using the EventsToSkip … OK. data43.txt.2
>> Tue Nov 3 13:48:19 2009 Run: 221295 Event: 19061 Stop: event << %% NOTICE-Processor.SkipBadEventsProc: Skipping event
Proceed on the rest of runs.
Use Skip Bad Event
Read the /home/dskim/DSkimPackage/tcl/skimD_SkipBadEventsProc.tcl
proc sel SkipBadEventsProc param SkipBadEventsProc EventsToSkip "/home/dskim/DSkimPackage/tcl/EventsToSkip.txt"
Add lines in loadHadronicDNtupleProc.tcl after the SkipBadRunsProc:
# Skip events which consum too much CPU time proc sel SkipBadEventsProc param SkipBadEventsProc EventsToSkip "/home/xs32/work/CLEO/analysis/DHad/src/2.2/gen/EventsToSkip.txt"
Add events in EventsToSkip.txt… works.
Proceed on the rest of the data43
From 221495 to 221898 … done. data43-test-1.txt
qsub dtuple-data43-test.sh
Run: 221504 Event: 5468 … 16 min. Run: 221699 Event: 31675 … 8 min. Run: 221705 Event: 29678 … 3:10 ! Run: 221715 Event: 65978 … 23 min.
From 221899 to 223271 … done. data43-test-2.txt
qsub dtuple-data43-test2.sh
Run: 221929 Event: 16197 … 8 min. Run: 221963 Event: 7013 … 17 min. Run: 221963 Event: 38551 … 4 min. Run: 221988 Event: 23888 … 22 min. Run: 222014 Event: 47972 … 3 min Run: 222019 Event: 62955 … 5 min. Run: 222131 Event: 8240 … 1hour! Run: 222153 Event: 69636 … 8 min. Run: 222207 Event: 32078 … 6 min. Run: 222279 Event: 65673 … 1:36 hour! Run: 222515 Event: 60769 … 6 min. Run: 222544 Event: 42858 … 38 min. Run: 222844 Event: 46989 … 12 min. Run: 223051 Event: 28048 … 17 min.
Skip events > 15 min…
| Run | Event | Time |
|---|---|---|
| 221242 | 6470 | 1:30 |
| 221295 | 19061 | 1:00 |
| 221493 | 61358 | 1:30 |
| 221705 | 29678 | 3:10 |
| 221715 | 65978 | 0:23 |
| 221963 | 7013 | 0:17 |
| 221988 | 23888 | 0:22 |
| 222131 | 8240 | 1:00 |
| 222279 | 65673 | 1:36 |
| 222544 | 42858 | 0:38 |
| 223051 | 28048 | 0:17 |
Clean the data43 … OK.
qsub dtuple-data43.sh
Job finished:
Job 1579825 (dtuple-data43.sh) Complete User = xs32 Queue = all.q@lnx187.lns.cornell.edu Host = lnx187.lns.cornell.edu Start Time = 11/04/2009 09:55:51 End Time = 11/05/2009 05:48:58 User Time = 16:01:55 System Time = 00:15:03 Wallclock Time = 19:53:07 CPU = 16:16:58 Max vmem = 1.848G Exit Status = 0
Output root files:
-rw-r--r-- 1 xs32 cms 941M Nov 5 05:48 data43_dskim_evtstore_1.root -rw-r--r-- 1 xs32 cms 1.8G Nov 4 23:49 data43_dskim_evtstore.root
Avoid the memeory leak for data43
Edit the code HadronicDNtupleProc.cc:
677 // *missingMassObjects.loosePi0List = TurnPhdPi0sIntoNavPi0ToGGs(loosePi0table, showtable); 678 // xs32: avoide the memory leak, only used for the missing mass calculation
1120 } // Added this xs32, ryd
| Run | Event | Time |
|---|---|---|
| 221242 | 6470 | 1:30 |
| 221705 | 29678 | 3:10 |
Test on run 221242,
Compile in 2.2 … OK.
Change the tcl file … OK.
Remove the event in the EventToSkip … OK.
qsub dtuple-data43-test.sh ...
Save log: data43-test.txt
Test on run 221705:
Change the tcl file … OK.
qsub dtuple-data43-test2.sh ...
Looks still wasting time on that … log: data43-test2.txt
| Run | Event | Time (old) | New |
|---|---|---|---|
| 221242 | 6470 | 1:30 | 1:30 |
| 221705 | 29678 | 3:10 | 3:33 |
Put Time Statement inside code
Ask Peter for advice … sent.
Proceed on data44, 45, 46
qsub dtuple-data44.sh qsub dtuple-data45.sh qsub dtuple-data46.sh
Data 44 job died:
Job 1579399 (dtuple-data44.sh) Aborted Exit Status = 152 Signal = XCPU User = xs32 Queue = all.q@lnx187.lns.cornell.edu Host = lnx187.lns.cornell.edu Start Time = 11/04/2009 09:25:06 End Time = 11/05/2009 02:55:35 CPU = 14:58:04 Max vmem = 1.869G failed assumedly after job because: job 1579399.1 died through signal XCPU (24)
Log file: dtuple-data44.txt
Output files:
-rw-r--r-- 1 xs32 cms 59M Nov 5 01:20 data44_dskim_evtstore_1.root -rw-r--r-- 1 xs32 cms 1.8G Nov 5 01:06 data44_dskim_evtstore.root
Need code to get the table of time consuming…
Data 45 job finished:
Job 1579400 (dtuple-data45.sh) Complete User = xs32 Queue = all.q@lnx189.lns.cornell.edu Host = lnx189.lns.cornell.edu Start Time = 11/04/2009 09:26:06 End Time = 11/04/2009 23:00:39 User Time = 12:33:38 System Time = 00:05:00 Wallclock Time = 13:34:33 CPU = 12:38:38 Max vmem = 1.743G Exit Status = 0
Output files:
-rw-r--r-- 1 xs32 cms 680M Nov 4 23:00 data45_dskim_evtstore_1.root -rw-r--r-- 1 xs32 cms 1.8G Nov 4 19:40 data45_dskim_evtstore.root
Data 46 job aborted:
Job 1579401 (dtuple-data46.sh) Aborted Exit Status = 152 Signal = XCPU User = xs32 Queue = all.q@lnx65107.lns.cornell.edu Host = lnx65107.lns.cornell.edu Start Time = 11/04/2009 09:26:35 End Time = 11/04/2009 20:46:50 CPU = 10:00:03 Max vmem = 1.863G failed assumedly after job because: job 1579401.1 died through signal XCPU (24)
Log file: dtuple-data46.txt
Output files:
-rw-r--r-- 1 xs32 cms 585M Nov 4 20:42 data46_dskim_evtstore_1.root -rw-r--r-- 1 xs32 cms 1.8G Nov 4 17:38 data46_dskim_evtstore.root
The possible solutions are :
- Break the job into two
- Skip the bad events (manual or with tools)
Process data44
Skip the bad events which take more than 20 minutes.
dhad-2.2 table time data44
| Run | Event | Duration |
|---|---|---|
| 223560 | 21396 | 1:16:05 |
| 223763 | 83953 | 0:27:06 |
| 223979 | 77317 | 0:26:26 |
| 224185 | 32495 | 0:34:37 |
| 224239 | 23780 | 0:29:34 |
| 224512 | 53492 | 1:04:57 |
| 224597 | 41817 | 0:28:44 |
It stopped at Run: 224597 Event: 46933, test this run …
Skip the 41817 .
qsub dtuple-data44-test.sh
Still stuck at this event… data44.txt
>> Sat Nov 14 11:29:37 2009 Run: 224597 Event: 46933 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of D candidates exceeded %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of DD candidates exceeded >> Sat Nov 14 14:43:29 2009 Run: 224597 Stop: none << %% INFO-JobControl.JobControl: Reached end of an active source %% INFO-JobControl.SourceManager: Removed source-in-use "eventstore"
This is a must skip event. Add in the EventsToSkip.txt. … OK.
Get the last run on the first root file: getMaxRun.C.
cd $dhad/dat/data/2.2
root -l
.L getMaxRun.C
getMaxRun("data44_dskim_evtstore.root")
Max Run: 224556
Use this run as the first run in the second job.
Find the starting run in DataSetSummary … 223279 - 226042
data44_1: 223279 224555 data44_2: 224556 226042
Submit jobs.
qsub dtuple-data44_1.sh qsub dtuple-data44_2.sh
Check the second job log:
>> Tue Nov 17 23:20:06 2009 Run: 225760 Event: 4270 Stop: event << %% NOTICE-Processor.RunEventNumberProc: Run: 225760 Event: 4270 Fill: Switching to new file: /home/xs32/work/CLEO/analysis/DHad/dat/data/2.2/data44_2_dskim_evtstore_1.root
Need to start the third job from run 225760.
Other long time runs …
cd $dhad/dat/data/2.2 dhad-2.2 table time dtuple-data44_2.txt
| Run | Event | Duration |
|---|---|---|
| 224597 | 46933 | 3:23:14 |
| 224650 | 8836 | 1:22:46 |
| 224664 | 48250 | 0:39:38 |
| 224668 | 19000 | 2:30:54 |
| 224689 | 70523 | 0:30:49 |
| 224765 | 62530 | 6:51:46 |
| 224867 | 44242 | 0:42:09 |
| 224898 | 33986 | 0:24:11 |
| 224907 | 8998 | 0:25:26 |
| 224909 | 10978 | 0:22:10 |
| 225080 | 69974 | 0:24:48 |
| 225519 | 76639 | 0:50:34 |
| 225599 | 57356 | 0:38:39 |
| 225665 | 34335 | 3:03:24 |
| 225718 | 92758 | 0:23:34 |
| 225764 | 13222 | 1:36:38 |
| 225785 | 15222 | 0:47:01 |
| 225878 | 57850 | 2:16:28 |
| 225991 | 19929 | 1:55:02 |
| 226031 | 963 | 0:28:26 |
Wait for finish then update the above table … done.
Update the EventsToSkip … done.
Update the tcl … done.
Update the sh … done.
Change the third file name:
mv data44_2_dskim_evtstore_1.root data44_3_dskim_evtstore.root
Process data46
Skip the bad events which take more than 20 minutes.
dhad-2.2 table time data46
| Run | Event | Duration |
|---|---|---|
| 228307 | 33006 | 1:06:27 |
| 228677 | 46521 | 0:27:29 |
| 229695 | 56504 | 0:29:09 |
The job stopped at Run: 229724 Event: 68512
Test on this run …
qsub dtuple-data46-test.sh
Log file data46.txt:
>> Sat Nov 14 11:42:04 2009 Run: 229724 Event: 68512 Stop: event << %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of D candidates exceeded %% NOTICE-Processor.HadronicDNtupleProc: Hard limit on number of DD candidates exceeded >> Sat Nov 14 13:51:31 2009 Run: 229724 Event: 69349 Stop: event << %% NOTICE-Processor.RunEventNumberProc: Run: 229724 Event: 69349 >> Sat Nov 14 13:51:34 2009 Run: 229724 Event: 78793 Stop: event << %% INFO-TrackManProd.TMData: The curvature of track 8 is 0. Setting the various radii to zero. >> Sat Nov 14 13:51:35 2009 Run: 229724 Stop: none << %% INFO-JobControl.JobControl: Reached end of an active source %% INFO-JobControl.SourceManager: Removed source-in-use "eventstore" Suez.loadHadronicDNtupleProc> exit
Spend 2 hours on this event. Skip it and the previous 3 events.
Break the job into two:
Get the last run number:
cd $dhad/dat/data/2.2
root -l
.L getMaxRun.C
getMaxRun("data46_dskim_evtstore.root")
Max Run: 229385
First job: 228285 229384 Second job: 229385 230130
qsub dtuple-data46_1.sh qsub dtuple-data46_2.sh
Check the first time table:
dhad-2.2 table time dtuple-data46_1.txt
| Run | Event | Duration |
|---|---|---|
| 228360 | 29626 | 0:28:20 |
| 229228 | 44793 | 0:30:28 |
| 229268 | 40581 | 0:21:13 |
dhad-2.2 table time dtuple-data46_2.txt
| Run | Event | Duration |
|---|---|---|
| 229632 | 75259 | 0:21:44 |
| 229653 | 72630 | 0:20:42 |
| 229778 | 69164 | 0:26:30 |
| 229800 | 39024 | 0:47:52 |
Update EventsToSkip … done.
Re-process data43
Update the skip events … OK.
qsub dtuple-data43.sh
Done.
Update the time table:
dhad-2.2 table time $dhad/dat/data/2.2/dtuple-data43.txt
| Run | Event | Duration |
|---|---|---|
| 221238 | 26193 | 0:37:26 |
Update this event to EventToSkip.txt … OK.
From the log file:
>> Tue Nov 17 01:02:30 2009 Run: 222627 Event: 29127 Stop: event << %% NOTICE-Processor.RunEventNumberProc: Run: 222627 Event: 29127 Fill: Switching to new file: /home/xs32/work/CLEO/analysis/DHad/dat/data/2.2/data43_dskim_evtstore_1.root
Confirm the number by script:
cd $dhad/dat/data/2.2
root -l
.L getMaxRun.C
getMaxRun("data43_dskim_evtstore.root")
Max Run: 222627
First job: 220898 222626 Second job: 222627 223271
Update the tcl file and sh file … OK.
Use them next time.
Change the file names:
mv data43_dskim_evtstore.root data43_1_dskim_evtstore.root mv data43_dskim_evtstore_1.root data43_2_dskim_evtstore.root
Re-process data45
Get the last run number:
cd $dhad/dat/data/2.2
root -l
.L getMaxRun.C
getMaxRun("data45_dskim_evtstore.root")
Max Run: 227841
Break job into two:
- 226429 227840
- 227841 228271
Edit the dataselection.tcl.
Get the bad events:
dhad-2.2 table time data45
| Run | Event | Duration |
|---|---|---|
| 226804 | 43667 | 0:24:34 |
| 227041 | 52921 | 0:30:03 |
| 227805 | 77572 | 0:21:16 |
| 227813 | 837 | 0:35:04 |
| 228247 | 28837 | 0:37:37 |
Update the EventToSkip… done.
qsub dtuple-data45_1.sh qsub dtuple-data45_2.sh
Update the time table … done.
dhad-2.2 table time dtuple-data45_1.txt dhad-2.2 table time dtuple-data45_2.txt
All skipped.
Remove the old root files:
rm data45_dskim_evtstore*.root
Extract Yields
Link the root files to 7.06/dat/data/537ipb
cd $dhad/7.06/dat/data ln -s $dhad/dat/data/2.2 537ipb
dhad-2.2 yield 537ipb -t d -m 0 OK.
dhad-2.2 yield 537ipb -t d -m 0 –qsub
Not work:
User = xs32 Queue = all.q@lnx326.lns.cornell.edu Host = lnx326.lns.cornell.edu Start Time = 11/19/2009 08:43:26 End Time = 11/19/2009 08:43:26 User Time = 00:00:00 System Time = 00:00:00 Wallclock Time = 00:00:00 CPU = NA Max vmem = NA Exit Status = 127
Check the log file:
/nfs/sge/root/default/spool/lnx326/job_scripts/1602642: line 36: dhad-2.2: command not found
Simple try:
dhad yield 537ipb -t d -m 1 –qsub
/nfs/sge/root/default/spool/lnx302/job_scripts/1602647: line 36: dhad: command not found
Add setdhad 2.2 in the sh file … OK.
dhad-2.2 yield 537ipb -t d -m 1 –qsub log-2009-11-19 10:26:54
dhad-2.2 yield 537ipb -t d -m 0 –qsub log-2009-11-19 10:31:11
dhad-2.2 yield 537ipb -t d -m 3 –qsub log-2009-11-19 10:31:36
dhad-2.2 yield 537ipb -t d -m 200 –qsub log-2009-11-19 10:31:43
dhad-2.2 yield 537ipb -t d -m 201 –qsub log-2009-11-19 10:31:48
dhad-2.2 yield 537ipb -t d -m 202 –qsub log-2009-11-19 10:31:55
dhad-2.2 yield 537ipb -t d -m 203 –qsub log-2009-11-19 10:32:01
dhad-2.2 yield 537ipb -t d -m 204 –qsub log-2009-11-19 10:32:07
dhad-2.2 yield 537ipb -t d -m 205 –qsub log-2009-11-19 10:32:13
Clean the duplicated files … done.
dhad yield 537ipb -t d -m 1 OK.
dhad-2.2 yield 537ipb -t d -m 0 –qsub log-2009-12-10 11:20:19
dhad-2.2 yield 537ipb -t d -m 1 –qsub log-2009-12-10 11:20:03
dhad-2.2 yield 537ipb -t d -m 3 –qsub log-2009-12-10 11:20:27
dhad-2.2 yield 537ipb -t d -m 200 –qsub log-2009-12-10 11:20:33
dhad-2.2 yield 537ipb -t d -m 201 –qsub log-2009-12-10 11:20:38
dhad-2.2 yield 537ipb -t d -m 202 –qsub log-2009-12-10 11:20:43
dhad-2.2 yield 537ipb -t d -m 203 –qsub log-2009-12-10 11:20:48
dhad-2.2 yield 537ipb -t d -m 204 –qsub log-2009-12-10 11:20:54
dhad-2.2 yield 537ipb -t d -m 205 –qsub log-2009-12-10 11:20:59
Do the Fits
setdhad 2.2 dhad-2.2 fit 537ipb -t d -m 0 OK.
dhad-2.2 fit 537ipb -t d -m 0 –qsub log-2009-11-23 15:29:29
dhad-2.2 fit 537ipb -t d -m 1 –qsub log-2009-11-23 15:29:39
dhad-2.2 fit 537ipb -t d -m 3 –qsub log-2009-11-23 15:29:47
dhad-2.2 fit 537ipb -t d -m 200 –qsub log-2009-11-23 15:29:52
dhad-2.2 fit 537ipb -t d -m 201 –qsub log-2009-11-23 15:29:57
dhad-2.2 fit 537ipb -t d -m 202 –qsub log-2009-11-23 15:30:02
dhad-2.2 fit 537ipb -t d -m 203 –qsub log-2009-11-23 15:30:07
dhad-2.2 fit 537ipb -t d -m 204 –qsub log-2009-11-23 15:30:12
dhad-2.2 fit 537ipb -t d -m 205 –qsub log-2009-11-23 15:30:17
After the clean up for the duplicated events, do the fit again:
dhad-2.2 fit 537ipb -t d -m 0 –qsub log-2009-12-10 14:12:28
dhad-2.2 fit 537ipb -t d -m 1 –qsub log-2009-12-10 14:12:33
dhad-2.2 fit 537ipb -t d -m 3 –qsub log-2009-12-10 14:12:42
dhad-2.2 fit 537ipb -t d -m 200 –qsub log-2009-12-10 14:12:49
dhad-2.2 fit 537ipb -t d -m 201 –qsub log-2009-12-10 14:12:54
dhad-2.2 fit 537ipb -t d -m 202 –qsub log-2009-12-10 14:12:59
dhad-2.2 fit 537ipb -t d -m 203 –qsub log-2009-12-10 14:13:04
dhad-2.2 fit 537ipb -t d -m 204 –qsub log-2009-12-10 14:13:09
dhad-2.2 fit 537ipb -t d -m 205 –qsub log-2009-12-10 14:13:14
Store the fitting plots
dhad-2.2 web plots 537ipb -t d
Extract yields for 818 data
Link the root files to 7.06/dat/data/537ipb
cd $dhad/7.06/dat/data mkdir 818ipb cd 818ipb
Fix the duplicated data45 files, changed the file names … done.
ln -s $dhad/dat/data/2.1/*.root . ln -s $dhad/dat/data/2.2/*.root .
dhad-2.2 yield 818ipb -t d -m 0 OK.
dhad-2.2 yield 818ipb -t d -m 0 –qsub log-2009-12-09 11:44:32
dhad-2.2 yield 818ipb -t d -m 1 –qsub log-2009-12-09 11:46:55
dhad-2.2 yield 818ipb -t d -m 3 –qsub log-2009-12-09 11:47:17
dhad-2.2 yield 818ipb -t d -m 200 –qsub log-2009-12-09 11:50:59
dhad-2.2 yield 818ipb -t d -m 201 –qsub log-2009-12-09 11:51:28
dhad-2.2 yield 818ipb -t d -m 202 –qsub log-2009-12-09 11:51:45
dhad-2.2 yield 818ipb -t d -m 203 –qsub log-2009-12-09 11:51:53
dhad-2.2 yield 818ipb -t d -m 204 –qsub log-2009-12-09 11:51:58
dhad-2.2 yield 818ipb -t d -m 205 –qsub log-2009-12-09 11:52:07
Fitting for 818 data
dhad-2.2 fit 818ipb -t d -m 0 ... OK.
dhad-2.2 fit 818ipb -t d -m 0 –qsub log-2009-12-09 15:06:18
dhad-2.2 fit 818ipb -t d -m 1 –qsub log-2009-12-09 15:07:05
dhad-2.2 fit 818ipb -t d -m 3 –qsub log-2009-12-09 15:07:14
dhad-2.2 fit 818ipb -t d -m 200 –qsub log-2009-12-09 15:07:21
dhad-2.2 fit 818ipb -t d -m 201 –qsub log-2009-12-09 15:07:26
dhad-2.2 fit 818ipb -t d -m 202 –qsub log-2009-12-09 15:07:31
dhad-2.2 fit 818ipb -t d -m 203 –qsub log-2009-12-09 15:07:36
dhad-2.2 fit 818ipb -t d -m 204 –qsub log-2009-12-09 15:07:41
dhad-2.2 fit 818ipb -t d -m 205 –qsub log-2009-12-09 15:07:46
Store fitting plots
dhad-2.2 web plots 818ipb -t d
Compare the yields
Refer to 200607 report table:
| Category | Y(281)/Y(56) |
|---|---|
| D0 ST | 5.00 ± 0.02 |
| D+ ST | 5.02 ± 0.03 |
| D0D0B DT | 5.00 ± 0.10 |
| D+ D- DT | 5.06 ± 0.12 |
| 281/55.8 | 5.04 |
And the efficiency table:
| Category | Y(281)/Y(56) | Y(281)/Y(56) (old) |
|---|---|---|
| D all ST | 5.16 ± 0.08 | 5.00 ± 0.02 |
| - | - | 5.02 ± 0.03 |
| D0 D0B DT | 4.90 ± 0.11 | 5.00 ± 0.10 |
| D+ D- DT | 4.83 ± 0.14 | 5.06 ± 0.12 |
| 281/55.8 | 5.04 | 5.04 |
Need to do the fits for 818. … done.
Compare the yields with 281 …
dhad-2.2 table compare yields data divide 281ipb 818ipb
Compare the 537 yields with 281 …
dhad-2.2 table compare yields data divide 281ipb 537ipb
Update for the error … OK.
Need to talk with David for about the 818 case where the new data was included…
Compare the fitting parameters
Compare one parameter first:
dhad-2.2 table compare parameter data sigmap1 281ipb 537ipb --set rnd=0.000001 ... OK
Compare the others … OK.
Process 281 data with eventstore 20070822 (2.2)
Data 31
Check the sh file dtuple-data31.sh … OK.
Change the dataselection.tcl :
if { ( $env(INPUTDATA) == "data31_dskim_evtstore" ) } {
# eventstore in 20050429 dtag all dataset data31
eventstore in 20070822 dtag all dataset data31
qsub dtuple-data31.sh … No output due to the later timestamp.
Stop all the jobs … OK.
Data 32
Change the file dtuple-data32.sh … OK.
Change the dataselection.tcl :
if { ( $env(INPUTDATA) == "data32_dskim_evtstore" ) } {
# eventstore in 20050429 dtag all dataset data32
eventstore in 20070822 dtag all dataset data32
Also change the data33, data35, data36, data37… OK.
qsub dtuple-data32.sh …
Data33
Change the file dtuple-data33.sh … OK.
qsub dtuple-data33.sh …
Data35
Change the file dtuple-data35.sh … OK.
qsub dtuple-data35.sh …
Data36
Change the file dtuple-data36.sh … OK.
qsub dtuple-data36.sh …
Data37
Change the file dtuple-data37.sh … OK.
qsub dtuple-data37.sh …
Prepare four-vector for CLEOG (2.3)
Generate
Start from dhad 2.1
cd $dhad/src cp -r 2.1 2.3 cd 2.3/python cvs tag -r V02-01-02 -b B02-03 cvs up -r B02-03
- Remove the rel in setup.sh
- Edit the DHad.py src=2.3
-
Edit fixmcjob.sh
Suggestion from Laura, check the HN from Indiana group.
Found page: https://wiki.lepp.cornell.edu/lepp/bin/view/CLEO/Private/SW/GeneratingMCDecayTree
-
Create gendecaytree.tcl
set fileout $env(OUTDIR)/cleog_decaytree_$env(DECAYTITLE)_$env(BATCH).pds
-
Edit cleog-generic-array.sh
suez -f gen_decaytree.tcl
-
Test for one mode for 10 events
setdhad 2.3
Edit cleog-generic-array.sh:
export NUMEVT=10
Test
dhad-2.3 gen cleog Single_Dm_to_Kspi task 1
Error: => cleog.txt
can't read "env(MCRUN)": no such variable
Change to RUNNUMBER. … => cleog.txt.1 (this file corrupts by a later call) stops.
Ask on HN… sent.
Message from Surik,
Suggestion from Dan, delete the commented line … => cleog.txt.2
can't read "env(USERDEC)": no such variable
Change the USERDEC to UDECAY … => cleog.txt.3
Try large number …
Edit cleog-generic-array.sh:
export NUMEVT=`cat $SCRIPTDIR/tag_numbers/$1`
Run … => cleog.txt.4 OK. Output pdf 5.5M.
Need mass production … hold.
Try to continue cleog in this release.
Re-name the process as 'decaytree':
Edit fixmcjob.sh.template.
Create decaytree-generic-array.sh.
Test 10 events.
dhad-2.3 gen decaytree Single_Dm_to_Kspi task 1
Save log file => decaytree.txt
Continue cleog in this release.
-
Try to remove the lines in the decay tree gen stage:
file out $fileout "beginrun {}" "startrun {}" "endrun {}" physics "event $treeMC"dhad-2.3 gen decaytree Single_Dm_to_Kspi task 2
Save log => decaytree.txt.1
No output.
Remove the line: => decaytree.txt.2
sink rm $fileout;
Has output.
Consult with Dan, the syntax is correct.
-
Generate one normal number mode
Edit decaytree-generic-array.sh:
export NUMEVT=`cat $SCRIPTDIR/tag_numbers/$1`
Restore gendecaytree.tcl:
file out $fileout "beginrun {}" "startrun {}" "endrun {}" physics "event $treeMC"dhad-2.3 gen decaytree Single_Dm_to_Kspi task 3
Output 5.5M.
Continue test in 9.03 … OK.
Clean up files … done.
-
Generate all modes
dhad-2.3 gen decaytree single ... problem.
Move to dhad-3.0.5 to deal with this.
Use four-vector for CLEOG
Edit the cleog-generic-array.sh.
Add $INDIR.
Create cleogdecaytree.tcl.
Test 10 events.
dhad-2.3 gen cleog Single_Dm_to_Kspi task 1
Save log file => cleog.txt
Same error.
Reply to Surik and HN …
Try the removed pds file task 2: => cleog.txt.1
dhad-2.3 gen cleog Single_Dm_to_Kspi task 2
Same error:
can't read "argArray(generatorProd)": no such element in array
Fix the bug of cleogcommand.tcl by Dan:
~dsr/clego_command.tcl
Edit line in cleogdecaytree.tcl :
#run_file $env(C3_SCRIPTS)/cleog_command.tcl run_file /home/dsr/cleog_command.tcl
Test 10 events.
dhad-2.3 gen cleog Single_Dm_to_Kspi task 1
Other error => cleog.txt.2
unknown cleoc pass2 tag "6" requested
Try it in the 9.03 … OK.
So, use this only at 9.03 for now.
Close this section.
Leave 9.03 for continue cleog and pass2.
Debug in the BeamEnergyProxy (2.4)
Setup 2.4
Based on 2.1.
cd $dhad/src cp -r 2.1 2.4 cp /nfs/cor/an1/ponyisi/2007-4/local_setup /home/xs32/work/CLEO/analysis/DHad/src/2.4/cleo
Backup the file. localsetup.peter
Change local_setup to use the local build.
Change the cleog-generic-array.sh … OK.
Change the python code DHad.py for 2.4 … OK.
Change the setup.sh … OK.
Change the fixmcjob.sh.template …
Test for 10 events …
dhad-2.4 gen cleog Single_Dm_to_Kpipipi0 task 3
OK. cleog.txt
Check out the CesrBeamEnergyPord
Look up the pkg versions for the CesrBeamEnergyProd
/nfs/cleo3/Offline/rel/20050525_MCGEN/pkg_versions
Found:
CesrBeamEnergyProd Infrastructure v01_00_02 proc
Check out the CesrBeamEnergyProd:
cd $dhad/src/2.4/cleo/ export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co -r v01_00_02 CesrBeamEnergyProd . local_setup c3make
Edit the BeamEnergyProxy.cc:
std::cout << "xs23_debug >>> beam energy:" << p_BeamEnergy.value() << std::endl;
Test for 10 events :
dhad-2.4 gen cleog Single_Dm_to_Kpipipi0 task 3
cleog.txt.1 no debug info.
But it is using the CesrBeamEnergyProd:
CesrBeamEnergyProd : /home/xs32/work/CLEO/analysis/DHad/src/2.4/cleo/build/Linux/shlib/CesrBeamEnergyProd.so_20050525_MCGEN
Suggestion from Anders: Add print statement in the beginning…
Try the report (INFO) …
/home/xs32/work/CLEO/analysis/DHad/src/2.4/cleo/CesrBeamEnergyProd/Class/BeamEnergyProxy.cc:157: no matching function for call to `std::auto_ptr<BeamEnergy>::value()' /home/xs32/work/CLEO/analysis/DHad/src/2.4/cleo/CesrBeamEnergyProd/Class/BeamEnergyProxy.cc:160: no matching function for call to `std::auto_ptr<BeamEnergy>::value()'
Check the method, put this before the reset.
std::cout << "xs23_debug >>> cesr beam energy:" << cesr->value() << std::endl;
Test …
dhad-2.4 gen cleog Single_Dm_to_Kpipipi0 task 3
std::cout << "xs23_debug >>> beam energy:" << p_BeamEnergy->value() << std::endl;
Test … no output.
Use the INFO.
report( INFO, kFacilityString )
<< "xs23_debug >>> beam energy:" << p_BeamEnergy->value() << endl ;
Set the DEBUG level in the tcl … OK.
Clean the previous log.
dhad-2.4 gen cleog Single_Dm_to_Kpipipi0 task 3
xs23!! -> Change to xshi! Std works too.
Need more digits:
#include <iostream>
#include <iomanip>
std::cout << "xshi_debug >>> beam energy:" << std::setprecision(8)
<< p_BeamEnergy->value() << std::endl;
Test …
xshi_debug >>> beam energy:1.8863001
Now find the number in the new release.
More precision in MCBeamParametersProxy
Look up the pkg versions for the MCSymmetricBeamProd
/nfs/cleo3/Offline/rel/20050525_MCGEN/pkg_versions
MCSymmetricBeamProd MonteCarlo v01_02_03 proc
Check out the code:
cd $dhad/src/2.4/cleo/ export CVSROOT=/nfs/cleo3/cvsroot cleo3cvs co -r v01_02_03 MCSymmetricBeamProd . local_setup c3make
Add precision in the code:
#include <iomanip> report( INFO, kFacilityString ) << "Nominal beam energy = " << std::setprecision(8)
Back up the previous log.
Test on 10 events …
dhad-2.4 gen cleog Single_Dm_to_Kpipipi0 task 3
Save log cleog.txt.1
%% NOTICE-ConstantsPhase2Delivery.DBCP2Proxy: DBBeamEnergyShift using version 1.27 %% DEBUG-CesrBeamEnergyProd.BeamEnergyProxy: Applying Beam Energy shift of -0.00102 GeV xshi_debug >>> beam energy:1.8863001 %% DEBUG-ConstantsPhase2Delivery.DBCP2Proxy: getConstants: returned successfully. %% NOTICE-ConstantsPhase2Delivery.DBCP2Proxy: DBBeamEnergySpread using version 1.11 %% INFO-MCSymmetricBeamProd.MCBeamParametersProxy: Nominal beam energy = 1.8863001, spread = 0.0015, min=3.7619936, max=3.7832068
Fit 537 data with new parameters (2.5)
Setup 2.5
cd $dhad/src mkdir 2.5 cd 2.5 cvs co -d python -r V02-02-02 dhad/src/python mkdir bash cp ../2.2/bash/setup.sh bash ln -s /home/xs32/work/CLEO/analysis/DHad/lib/python/hep python
Change the src in DHad.py to 2.5.
cd $dhad/7.06/dat/evt ln -s 537ipb 537ipbv5
Test:
dhad-2.5 fit 537ipbv5 -t d -m 0 ... OK.
Change the parameters
Mass = 3.7718 GeV => 3.7724 Width = 0.0286 GeV => 0.0252 R = 12.3 => 12.7
dhad-2.5 fit 537ipbv5 -t d -m 0 ... OK.
Do the fits
dhad-2.5 fit 537ipbv5 -t d -m 0 –qsub log-2010-03-10 11:51:11
dhad-2.5 fit 537ipbv5 -t d -m 1 –qsub log-2010-03-10 11:52:15
dhad-2.5 fit 537ipbv5 -t d -m 3 –qsub log-2010-03-10 11:52:22
dhad-2.5 fit 537ipbv5 -t d -m 200 –qsub log-2010-03-10 11:52:27
dhad-2.5 fit 537ipbv5 -t d -m 201 –qsub log-2010-03-10 11:52:32
dhad-2.5 fit 537ipbv5 -t d -m 202 –qsub log-2010-03-10 11:52:38
dhad-2.5 fit 537ipbv5 -t d -m 203 –qsub log-2010-03-10 11:52:43
dhad-2.5 fit 537ipbv5 -t d -m 204 –qsub log-2010-03-10 11:52:48
dhad-2.5 fit 537ipbv5 -t d -m 205 –qsub log-2010-03-10 11:52:53
Fits are all OK.
Save the plots … figure
dhad-2.5 web plots 537ipbv5 -t d
Date: 2010-03-10 16:34:47 EST